Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Agriculture, Dairy & Animal Science
Eukaryotic organelles supposedly evolved from their bacterial ancestors because of their benefits to host cells. However, organelles are quite often retained, even when the beneficial metabolic pathway is lost, due to something other than the original beneficial function. The organellar function essential for cell survival is, in the end, the result of organellar evolution, particularly losses of redundant metabolic pathways present in both the host and endosymbiont, followed by a gradual distribution of metabolic functions between the organelle and host.
- organelle
- endosymbiosis
- plastid
- mitochondrion
- benefit
1. Introduction
A eukaryotic cell is typical by hosting semiautonomous organelles, such as mitochondria and plastids. These organelles are deeply integrated into the host cell; however, they usually keep some level of independence by encoding a fraction of the organellar proteome and RNAs in their genomes [1][2][3][4], living to some extent like endosymbiotic bacteria [5][6]. Mitochondria and plastids are, with few exceptions, essential for the host cell survival; once the cell has captured an organelle, it can hardly get rid of it [1][2][3][4][6][7]. It is believed that mitochondria and plastids evolved in endosymbiotic events, involving an engulfment or invasion of a free-living organellar ancestor, followed by the endosymbiotic transfer of genes from the captured entity to the nucleus of the host cell, with a consequent import of nuclear-encoded proteins into the organelle [3][8][9]. Symbiosis is an intimate, long-time relationship of two dissimilar organisms living together [10]. Although it is often understood as mutualism, the state beneficial for both partners, symbiosis, in fact, involves a continuum of relationships ranging from mutualism to parasitism [11]. The evolutionary history of plastids by domesticating a cyanobacterium is apparent because they are evolutionarily younger, and a cyanobacterial ancestor was likely acquired by the regular eukaryotic cell capable of phagocytosis [3][8][9].
2. The Plastid Benefit for the Host Is Photosynthesis
When talking about organelles, it is believed that benefits drive symbiotic relationships. However, it can be pretty challenging to trace the original benefit for which the endosymbiont is retained and integrated into the host cell. It is most likely that photosynthesis was the reason for adopting a cyanobacterial symbiont by a heterotrophic eukaryotic host (Figure 1) because there is no other way to get photosynthesis into the eukaryotic cell lacking it. Photosynthesis is managed by such complex molecular machinery that convergent evolution of the machinery appears highly improbable [6]. While the lateral gene transfer (LGT) likely stands behind the evolution of photosynthesis in prokaryotes [12][13][14], it played, except the endosymbiotic LGT, no role in the evolution of photosynthesis in eukaryotes. Instead, the evolutionary history of eukaryotic phototrophs is full of endosymbiotic events involving prokaryotic (at least twice) or eukaryotic phototrophs (many times) as donors of photosynthetic ability. Consequently, compartmentalization always physically separates photosynthesis from the host cell in eukaryotes (Figure 1). Photosynthesis has been transmitted throughout the tree of life for the apparent reason of the acquisition of a photoautotrophic lifestyle. Although it was supposed for a long time that plastid endosymbiotic events are rare in evolution [15][16], it recently appeared that at least two and six independent events were likely responsible for the appearance of primary and complex plastids, respectively, not counting complex plastids replacements. In addition to the primary endosymbiotic Archaeplastida, a relatively recent event involving heterotrophic amoebae and a cyanobacterium was proposed for the rhizarian Paulinella chromatophora, again with the apparent benefit of photoautotrophy [17][18][19][20][21]. Complex plastids have likely been independently acquired in Euglenophyta, Chorarachniophyta, Alveolata, Stramenopila, Haptophyta, and Cryptophyta [22][23][24][25][26][27][28] (Table 1).
Table 1. Selected plastids and their characteristics in various eukaryotes. It demonstrates the reductive evolution of plastids in eukaryotes.
| Organism | Supergroup | Type of the Plastid | Genes (Cds) | Genome | Reference |
|---|---|---|---|---|---|
| Arabidopsis thaliana | Archaeplastida | primary | 85 | circular | [29] |
| Porphiridium purpureum | Archaeplastida | primary | 224 | circular | [30] |
| Helicosporidium sp. | Archaeplastida | primary | 26 | circular | [31] |
| Polytomella sp. | Archaeplastida | primary | 0 | circular | [32] |
| Paulinella chromatohpora | Cercozoa (SAR) | primary (cyanelle) | 867 | circular | [33] |
| Euglena gracilis | Eugenophyta | complex (secondary) | 67 | circular | [34] |
| Euglena longa | Eugenophyta | complex (secondary) | 46 | circular | [35] |
| Heterocapsa triquetra | Dinophyta (SAR) | complex | 14 | Circular (minicircles) | [36] |
| Hematodinium sp. | Dinophyta (SAR) | - | - | - | [37] |
| Thalassiosira pseudonana | Bacillariophyta (SAR) | complex | 141 | circular | [38] |
| Chromera velia | Apicomonada (SAR) | complex | 78 | linear | [39] |
| Vitrella brassicaformis | Apicomonada (SAR) | complex | 94 | circular | [39] |
| Toxoplasma gondii | Sporozoa (SAR) | complex | 29 | circular | NCBI |
| Cryptosoridium muris | Sporozoa (SAR) | - | - | - | [40] |
Photosynthesis as a beneficial function is, however, not essential for the host cell‘s survival. Many photoautotrophic lineages became secondarily heterotrophic (Figure 1 and Table 1). Various eukaryotes have lost photosynthesis, being either forced by the lack of access to light or by chemical (e.g., antibiotic) disruption of the photosynthetic molecular machine [11][28][41][42][43][44][45][46][47]. For example, apicomplexan parasites (Sporozoa, Apicomplexa) likely became secondarily heterotrophic because of the easy availability of the organic carbon from the host or by the switch of the ancestral photoparasite from translucent to the opaque host [11]. Additionally, we have to consider that photosynthesis is not only beneficial for the primary producer, as it is quite costly and stands behind the outstanding production of reactive oxygen species (ROS), which can heavily damage the cell [42]. Additionally, a strictly phototrophic lifestyle forces the organisms to live in the access to light and thus limits their environmental distribution.
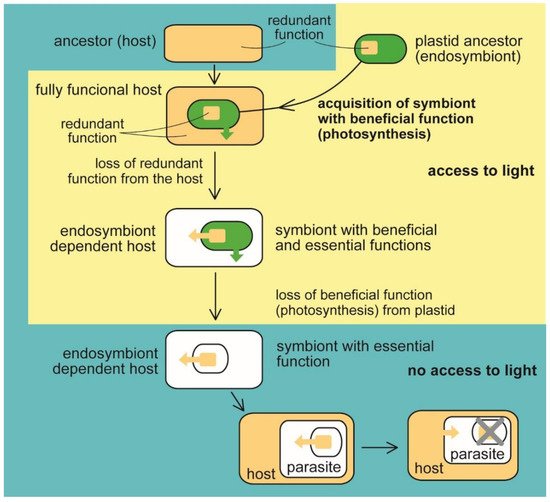
Figure 1. Evolution of benefit and essential function in the plastid. The heterotrophic host acquired a photosynthetic endosymbiotic bacterium with the function (photosynthesis) beneficial for the host. The host cell lost the redundant function (e.g., synthesis of heme, fatty acids, and isoprenoids). At the same time, the delegation of the syntheses to the endosymbiont makes it essential for host survival (eukaryotic phototrophs, e.g., Archaeplastida and Paulinella sp., and algae with complex plastids such as Ochrophyta, Cryptophyta, Haptophyta, Dinophyta, Apicomplexa, Euglenopyhta, Chlorarachniophyta [1][2][3][11]). The endosymbiont retained its indispensability for the host even when it had lost photosynthesis, the original beneficial function (in nonphotosynthetic algae, e.g., Helicosporidium sp., Polytomella sp., Euglena longa, apicomplexan parasites, for example, Plasmodium falciparum, Toxoplasma gondii [1][2][3][7][11] Table 1). Switching to parasitism and scavenging the essential compounds from the host allows the complete loss of the plastid (apicomplexan parasite Cryptosporidium [39], parasitic dinoflagellate Hematodinium [37]).
Therefore, many phototrophs are, in fact, mixotrophs, which can still live heterotrophically. Such organisms may be prone to losing photosynthesis when getting to the nutrient-rich environment or finding a successful heterotrophic strategy, such as predation or parasitism. Moreover, many protists combine the phototrophic and heterotrophic lives to overcome a reduced availability of various compounds present in host or prey but rare in their environment, such as, for example, nitrogen, phosphorus, iron, and sulfur [11]. Therefore, photosynthesis was frequently lost from green parasitic algae [28][41], euglenophytes [43], apicomplexan parasites [11][28][44][45][46][47], and dinoflagellates [45]. Various secondarily heterotrophic strategies, including parasitism, have evolved repeatedly during the evolution of life among former phototrophs [11][35][36]. It is worth noting that such trophic switches are often found in the same taxonomic groups, such as, for example, in myozozans, group of alveolate protists, consisting of dinoflagellates, apicomplexan parasites, and apicomonads (chromerids and colpodellids). Apicomplexans and dinoflagellates are also the only known algae shown to lose their plastids completely. The plastid is absent from the apicomplexan parasitic genus Cryptosporidium [39] and gregarines [48], and the parasitic dinoflagellates of the genus Hematodinium [37]. The plastid losses have happened exclusively in parasites thanks to their ability to scavenge the essential compounds originally produced by plastids from the host (Table 2).
Table 2. Examples of mitochondria and MROs and their characteristics in various eukaryotes. It demonstrates the reductive evolution of mitochondria, from mitochondrial organelles with large genomes to hydrogenosomes and mitosomes lacking genome and eukaryotic cells without mitochondrion.
| Species | Taxonomy | Type of Mitochondrion | Genes (Cds) | Genome | Reference |
|---|---|---|---|---|---|
| Andalucia godoyi | Jakobida | Aerobic/OXPHOS | 67 | circular | [49] |
| Reclinomonas americana | Jakobida | Aerobic/OXPHOS | 66 | circular | [50] |
| Homo sapiens | Metazoa (Obazoa) | Aerobic/OXPHOS | 13 | circular | [51] |
| Nymphaea colorata | Archaeplastida | Aerobic/OXPHOS | 42 | circular | [52] |
| Nyctotherus ovalis | Ciliophora (SAR) | Anaerobic/H-producing | 16 | linear | [53] |
| Plasmodium falciparum | Sporozoa (SAR) | Aerobic/OXPHOS | 3 | linear | [54] |
| Chromera velia | Apicomonada (SAR) | Aerobic/OXPHOS | 2 | linear | [55] |
| Amebophrya ceratii | Dinophyta (SAR) | Aerobic/OXPHOS | 0 | - | [56] |
| Neocallimastix sp. | Chytridiomycota (Obazoa) | Hydrogenosome | - | - | [57] |
| Giardia intestinalis | Metamonada | Mitosome (Fe-S clusters) | - | - | [57] |
| Monocercomonoides sp. | Oxymonadida | - | - | - | [58] |
3. A Beneficial Function of the Mitochondrion
In contrast with plastids, the hypothetical benefit responsible for acquiring mitochondria is the subject of speculation. Frankly speaking, it is not even clear if the mitochondrion was indispensable for forming an early eukaryotic cell [59] or when it was acquired in the course of evolution (early versus late acquisition). The discovery of the eukaryote lacking a mitochondrion proved that the organelle is not essential for the eukaryotic cell as it exists now when the organism is a secondary anaerobic (Figure 2) [58]. Generally, mitochondria are great examples of reductive evolution. The diversity of this organelle involves mitochondria with large circular genomes (e.g., in jakobids [49][50][60]), mitochondria with highly reduced genomes (e.g., those in apicomplexans; [61]), mitochondria-related organelles (MRO), e.g., mitosomes and hydrogenosomes without genomes [57], and, in the end, completely lost mitochondrion (in oxymonads; Table 2 [58]). Consequently, various such organelles possess diverse molecular machinery: the complete respiratory chain of aerobic mitochondria or variously reduced respiratory chains lacking particular complexes: complex I (myzozans, e.g., apicomplexan parasites, and some fungi), complexes III and IV, while complexes I, II, and V retained [62], complexes I and III in chromerids [55][61] and the dinoflagellates of the genus Amoebophrya [56]), or complexes III, IV, and ATP synthase in (some) hydrogenosomes [57]. The missing complexes are substituted by alternative sources of electrons (e.g., alternative NADH dehydrogenases, L- and D- lactate cytochrome c oxidoreductases), or the electron transport chain was even completely lost (MRO).
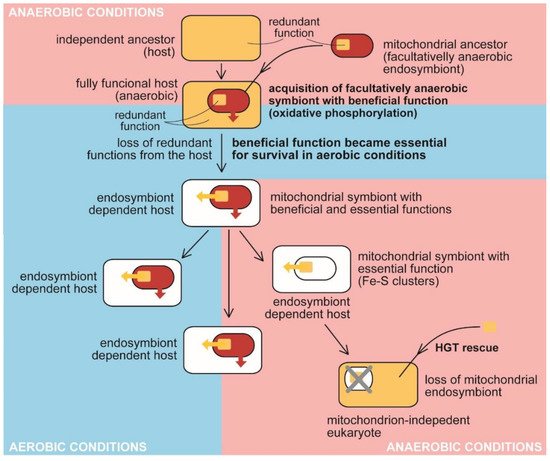
Figure 2. The traditional view on the evolution of benefit and essential function in the mitochondrion. Anaerobic host acquired a facultatively anaerobic endosymbiotic bacterium with the function beneficial for the host, presumably detoxifying oxygen. It became essential for the host in aerobic conditions. The redundant function was lost from the host (e.g., synthesis and assembly of Fe-S clusters). At the same time, the delegation of the synthesis to the endosymbiont makes it essential for host survival. The endosymbiont retained its indispensability for the host even when it had lost the original beneficial function by adaptating to anaerobic conditions. The acquisition of bacterial Fe-S clusters synthesis and assembly in the cytosol of oxymonads through HGT allowed the loss of the mitochondrion (MRO) [58].
Mitochondria host a wide range of metabolic functions, with oxidative phosphorylation being the most prominent (Figure 2), although it is found only in classical mitochondrial organelles, and lost from highly reduced MROs. In addition to that, mitochondria can be responsible for the metabolism of amino acids and nucleotides, steroid biosynthesis, heme synthesis, fatty acid catabolism, iron-cluster biogenesis, and many others [57][63][64][65][66]. Such metabolic complexity makes the search for the original beneficial function of the mitochondrion difficult. Various hypotheses have been formulated to explain the primary reason for acquiring a mitochondrion. It is further complicated by extensive genetic rearrangements of the organelle during organellogenesis because a substantial part of the mitochondrial proteome does not originate from the supposed mitochondrial ancestor [57].
The hydrogen hypothesis [67] proposed that the primary benefit of pre-mitochondrial symbiont was hydrogen production for the host, methanogenic Archaea. Some eukaryotes, such as Acanthamoeba castellanii (Amoebozoa), Brevimastigomonas motovehiculus (Rhizaria), Blastocystis spp. (Stramenopila), Nyctotherus ovalis (Alveolata) still contain hydrogen-producing mitochondria with complete (Acanthamoeba) or reduced respiratory chains [57][62][68]. Others host hydrogenosomes, organelles believed to represent modified hydrogen-producing mitochondria [69][57], which have lost the ATP generating part of the respiratory chain and retain just complex I (NADH hydrogenase), or complex II (succinate dehydrogenase) or both [53]. Martin et al. [69] also claim that the mitochondrial ancestor was a facultatively anaerobic bacterium.
Another possible beneficial function of a pre-mitochondrion was proposed by Thomas Cavalier-Smith [70], who speculated that the ancestor of mitochondrion was a photosynthetic purple bacterium, and the primary benefit was photoautotrophy, similar to plastids. This hypothesis assumes that both symbiont and host were facultative aerobes and that the host already has oxidative phosphorylation. A phototrophic symbiont would have an immediate intracellular synergy between a photosynthetic symbiont fixing CO2 and respiring and a phagotrophic host using oxygen and excreting CO2 [70].
Other hypotheses suppose that the primary benefit of the mitochondrion is related to dealing with free oxygen in the environment, preferring aerobic heterotrophic respiring bacterium as a mitochondrial ancestor [57]. Oxygen appeared in higher levels during the Great Oxidation Event between 2.4 and 2.1 Bya (billion years ago) [71]. If we take into account the possibility of early acquisition of mitochondrion, the oldest estimates of its appearance touch 2.1 Bya (1655–2094 Mya), while the youngest move around 1 Bya (943–1102 Mya) [72]. One of the earliest views on mitochondrial evolution supposes that the mitochondrion-free anaerobic eukaryotic ancestor acquired aerobic mitochondrion to detoxify oxygen accumulated in the environment after the Great Oxidation Event [73][74]. This scenario would even fit the timing of appearance of eukaryotes between 1 and 2 Bya. However, geochemical data indicate relatively low oxygen levels during the diversification of eukaryotes [71][75]. Moreover, Zimorski et al. [75] argued that those are metabolic processes in the mitochondrion, particularly electron transport chain generating reactive oxygen species, which may harm the cell and must be detoxicated. Therefore, oxidative phosphorylation can hardly be the primary benefit, at least in light of the oxygen detoxification hypothesis. However, we cannot ignore the fact that there is no eukaryote without classical mitochondrion with oxidative phosphorylation known that can permanently live in the presence of oxygen. The use of enzymes that can react with free oxygen does not allow obligate anaerobes to inhabit an oxygen environment. When looking at strictly anaerobic bacteria, they rely on low-potential flavoproteins used for anaerobic respiration. Exposure to oxygen likely causes superoxide and hydrogen peroxide production. They inactivate enzymes with these functional groups through the oxidation of dehydratase iron-sulfur clusters and sulphydryls. However, anaerobes utilize several classes of dioxygen-sensitive enzymes absent from aerobes, which maintain the redox balance during anaerobic fermentation. Their reaction mechanisms require exposure of the solvent of radicals or low-potential metal clusters to the oxygen that can react with it. Additionally, hydroxyl radicals damaging DNA and other biomolecules are generated because hydrogen peroxide oxidizes free iron [76]. Analogously, we can expect that oxygen could not be tolerated by anaerobes involved as the host cell in the early eukaryotic endosymbiotic events. A mitochondrial ancestor could eventually invade the host cell as an energy parasite. The proposed intracellularly parasitic ancestor of the mitochondrion was predicted to bear the ATP/ADP translocase importing ATP from the host. In such a case, there was no beneficial function behind a selection of an endosymbiont (parasite) in the event because the benefit was provided by the host cell [77].
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms10010122
References
- Gould, S.B.; Waller, R.F.; McFadden, G.I. Plastid evolution. Annu. Rev. Plant. Biol. 2008, 59, 491–517.
- Keeling, P.J. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Ann. Rev. Plant. Biol. 2013, 64, 583–607.
- Archibald, J.M. Endosymbiosis and eukaryotic cell evolution. Curr. Biol. 2015, 25, R911–R921.
- Gruber, A. What’s a name? Why organelles of endosymbiotic origin are implicitly called by their eukaryotic species name and how they can be distinguished from endosymbionts. Microb. Cell 2019, 6, 123–133.
- Pallen, M.J. Time to recognize that mitochondria are bacteria? Trends Microbiol. 2011, 19, 58–61.
- Oborník, M. In the beginning was the word: How terminology drives our understanding of endosymbiotic organelles. Microb. Cell 2019, 6, 134–141.
- Oborník, M. Endosymbiotic evolution of algae, secondary heterotrophy and parasitism. Biomolecules 2019, 9, 266.
- Martin, W.; Rujan, T.; Richly, E.; Hansen, A.; Cornelsen, S.; Lins, T.; Leister, D.; Stoebe, B.; Hasegawa, M.; Penny, D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA 2002, 99, 12246–12251.
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135.
- Keeling, P.J.; McCutcheon, J.P. Endosymbiosis: The feeling is not mutual. J. Theor. Biol. 2017, 434, 75–79.
- Oborník, M. Photoparasitism as an intermediate state in the evolution of apicomplexan parasites. Trends Parasitol. 2020, 36, 727–734.
- Raymond, J. The role of horizontal gene transfer in photosynthesis, oxygen production, and oxygen tolerance. Methods Mol. Biol. 2009, 532, 323–338.
- Koblížek, M.; Moulisová, V.; Muroňová, M.; Oborník, M. Horizontal transfers of two types of puf operons among phototrophic members of the Roseobacter clade. Folia Microbiol. 2015, 60, 37–43.
- Fischer, W.W.; Hemp, J.; Johnson, J.E. Evolution of oxygenic photosynthesis. Annu. Rev. Earth Planet Sci. 2016, 44, 647–683.
- Cavalier-Smith, T. Principles of protein and lipid targeting in secondary symbiogenesis: Euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Euk. Microbiol. 1999, 46, 347–366.
- Cavalier-Smith, T. The phagotrophic origin of eukaryotes and phylogenetic classification of protozoa. Int. J. Syst. Evol. Microbiol. 2002, 52, 297–354.
- Delaye, L.; Valadez-Cano, C.; Perez-Zamorano, B. How really ancient is Paulinella chromatophora? PLoS Curr. Tree Life 2016, 8.
- Stephens, T.G.; Gabr, A.; Calatrava, V.; Grossman, A.R.; Bhattacharya, D. Why is primary endosymbiosis so rare? New Phytol. 2021, 231, 1693–1699.
- Kies, L. Electron microscopical investigations on Paulinella chromatophora Lauterborn, a thecamoeba containing blue-green endosymbionts (cyanelles). Protoplasma 1974, 80, 69–89.
- Kies, L.; Kremer, B.P. Function of cyanelles in the thecamoeba Paulinella chromatophora. Naturwissenschaften 1979, 66, 578–579.
- Marin, B.; Nowack, E.C.; Melkonian, M. A plastid in the making: Evidence for a second primary endosymbiosis. Protist 2005, 156, 425–432.
- Bodył, A.; Stiller, J.W.; Mackiewicz, P. Chromalveolate plastids: Direct sescent or multiple endosymbioses? Trend. Ecol. Evol. 2009, 24, 119–121.
- Stiller, J.W.; Schreiber, J.; Yue, J.; Guo, H.; Ding, Q.; Huang, J. The evolution of photosynthesis in chromist algae through serial endosymbioses. Nat. Commun. 2014, 5, 5764.
- Petersen, J.; Ludewig, A.K.; Michael, V.; Bunk, B.; Jarek, M.; Baurain, D.; Brinkman, H. Chromera velia, endosymbioses and the rhodoplex hypothesis—plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages). Genome Biol. Evol. 2014, 6, 666–684.
- Dorrell, R.G.; Bowler, C. Secondary plastids of stramenopiles. Adv. Bot. Res. 2017, 84, 57–103.
- Strassert, J.F.H.; Irisarri, I.; Williams, T.A.; Burki, F. A molecular timescale of eukaryote evolution with implications for the origin of red algal-derived plastids. Nat. Commun. 2021, 12, 1879.
- Oborník, M. The birth of red complex plastids: One, three, or four times? Trends Parasitol. 2018, 34, 923–925.
- Hadariová, L.; Vesteg, M.; Hampl, V.; Krajčoviš, J. Reductive evolution of chloroplasts in non-photosynthetic plants, algae and protists. Curr. Genet. 2018, 64, 365–387.
- Sato, S.; Nakamura, Y.; Kaneko, T.; Asamizu, E.; Tabata, S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999, 29, 283–290.
- Tajima, N.; Sato, S.; Maruyama, F.; Kurokawa, K.; Ohta, H.; Tabata, S.; Sekine, K.; Moriyama, T.; Sato, N. Analysis of the complete plastid genome of the unicellular red alga Porphyridium purpureum. J. Plant Res. 2014, 127, 389–397.
- De Koning, A.P.; Keeling, P.J. The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 2006, 4, 12.
- Smith, D.R.; Lee, R.W. A Plastid without a Genome: Evidence from the Nonphotosynthetic Green Algal Genus Polytomella. Plant Physiol. 2014, 164, 1812–1819.
- Nowack, E.C.M.; Melkonian, M.; Glockner, G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 2008, 18, 410–418.
- Hallick, R.B.; Hong, L.; Drager, R.G.; Favreau, M.R.; Monfort, A.; Orsat, B.; Spielmann, A.; Stutz, E. Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res. 1993, 21, 3537–3544.
- Gockel, G.; Hachtel, W. Complete gene map of the plastid genome of the nonphotosynthetic euglenoid flagellate Astasia longa. Protist 2000, 151, 347–351.
- Zhang, Z.; Green, B.R.; Cavalier-Smith, T. Single gene circles in dinoflagellate chloroplast genomes. Nature 1999, 400, 155–159.
- Gornik, S.G.; Cassin, A.M.; MacRae, J.I.; Ramprasad, A.; Rchiad, Z.; McConville, M.J.; Bacic, A.; McFadden, G.I.; Pain, A.; Waller, R.F. Endosymbiosis undone by stepwise elimination of the plastid in a parasitic dinoflagellate. Proc. Natl. Acad. Sci. USA 2015, 112, 5767–5772.
- Oudot-Le Secq, M.-P.; Grimwood, J.; Shapiro, H.; Armbrust, E.V.; Bowler, C.; Green, B.R. Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: Comparison with other plastid genomes of the red lineage. Mol. Genet. Genom. 2007, 277, 427–439.
- Janouškovec, J.; Horák, A.; Oborník, M.; Lukeš, J.; Keeling, P.J. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc. Natl. Acad. Sci. USA 2010, 107, 10949–10950.
- Zhu, G.; Marchewka, M.J.; Keithly, J.S. Cryptosoridium parvum appears to lack a plastid genome. Microbiology 2000, 146, 315–321.
- Suzuki, S.; Endoh, R.; Manabe, R.; Ohkuma, M.; Hirakawa, Y. Multiple losses of photosynthesis and convergent reductive genome evolution in the colourless green algae Prototheca. Sci. Rep. 2018, 8, 940.
- Khorobrykh, S.; Havurinne, V.; Mattila, H.; Tyystjärvi, E. Oxygen and ROSS in photosynthesis. Plants 2020, 9, 91.
- Füssy, Z.; Záhonová, K.; Tomčala, A.; Krajčovič, J.; Yurchenko, V.; Oborník, M.; Eliáš, M. The cryptic plastid of Euglena longa defines a new type of nonphotosynthetic plastid organelle. mSphere 2020, 5, e00675-20.
- Salomaki, E.D.; Kolisko, M. There is treasure everywhere: Reductive plastid evolution in Apicomplexa in light of their close relatives. Biomolecules 2019, 9, 378.
- Waller, R.F.; Kořený, L. Plastid complexity in dinoflagellates: A picture of gains, losses, replacements and revisions. Adv. Bot. Res. 2017, 84, 105–143.
- Mathur, V.; Kolísko, M.; Hehenberger, E.; Irwin, N.A.T.; Leander, B.S.; Kristmundsson, Á.; Freeman, M.A.; Keeling, P.J. Multiple independent origins of apicomplexan-like parasites. Curr. Biol. 2019, 29, 1–6.
- Janouškovec, J.; Paskerova, G.G.; Miroliubova, T.S.; Mikhailov, K.V.; Birley, T.; Aleoshin, V.V.; Simdyanov, T.G. Apicomplexan-like parasites are polyphyletic and widely but selectively dependent on cryptic plastid organelles. eLife 2019, 8, e49662.
- Toso, M.A.; Omoto, C.K. Gregarina niphandrodes may lack both a plastid genome and organelle. J. Eukaryot. Microbiol. 2007, 54, 66–72.
- Gray, W.; Burger, G.; Derelle, R.; Klimeš, V.; Leger, M.M.; Sarrasin, M.; Vlček, Č.; Roger, A.J.; Eliáš, M.; Lang, B.F. The draft nuclear genome sequence and predicted mitochondrial proteome of Andalucia godoi, a protist with the most gene-rich and bacteria-like mitochondrial genome. BMC Biol. 2020, 18, 22.
- Lang, B.F.; Burger, G.; O’Kelly, C.J.; Cedergren, R.; Golding, G.B.; Lemieux, C.; Sankoff, D.; Turmel, M.; Gray, M.W. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 1997, 387, 493–497.
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.L.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465.
- Dong, S.; Zhao, C.; Chen, F.; Liu, Y.; Zhang, S.; Wu, H.; Zhang, L.; Liu, Y. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genom. 2018, 19, 614.
- De Graaf, R.M.; Ricard, G.; van Alen, T.A.; Duarte, I.; Dutilh, B.E.; Burgtorf, C.; Kuiper, J.W.P.; van der Staay, G.W.M.; Tielens, A.G.M.; Huynen, M.A.; et al. The Organellar Genome and Metabolic Potential of the Hydrogen-Producing Mitochondrion of Nyctotherus ovalis. Mol. Biol. Evol. 2011, 28, 2379–2391.
- Gardner, M.J.; Bates, P.A.; Ling, I.T.; Moore, D.J.; McCready, S.; Gunasekera, M.B.R.; Wilson, R.J.M.; Williamson, D.H. Mitochondrial DNA of the human malarial parasite Plasmodium falciparum. Mol. Biochem. Prasitol. 1988, 31, 11–17.
- Flegontov, P.; Michálek, J.; Janouškovec, J.; Lai, D.H.; Jirků, M.; Hajdušková, E.; Tomčala, A.; Otto, D.T.; Keeling, P.J.; Pain, A.; et al. Divergent mitochondrial respiratory chains in phototrophic relatives of apicomplexan parasites. Mol. Biol. Evol. 2015, 32, 1115–1131.
- John, U.; Lu, Y.; Wohlrab, S.; Groth, M.; Janouškovec, J.; Kohli, G.S.; Mark, F.C.; Bickmeyer, U.; Frahat, S.; Felder, M.; et al. A aerobic eukaryotic parasite with functional mitochondria that likely lacks a mitochondrial genome. Sci. Adv. 2019, 5, 1110.
- Roger, A.J.; Muñoz-Gómez, S.A.; Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. 2017, 27, R1177–R1192.
- Karnkowska, A.; Vacek, V.; Zubacova, Z.; Treitli, S.C.; Petrzelková, R.; Eme, L.; Novaqk, L.; Zarsky, V.; Barlow, L.D.; Herman, E.K. A eukaryote without a mitochondrial organelle. Curr. Biol. 2016, 26, 1274–1284.
- Hampl, V.; Čepička, I.; Eliáš, M. Was the mitochondrion necessary to start eukaryogenesis? Trends Microbiol. 2018, 27, 96–104.
- Burger, G.; Gray, M.W.; Forget, L.; Lang, B.F. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genom. Biol. Evol. 2013, 5, 418–438.
- Oborník, M.; Lukeš, J. The organellar genomes of Chromera and Vitrella, the phototrophic relatives of apicomplexan parasites. Ann. Rev. Microbiol. 2015, 69, 129–144.
- Müller, M.; Mentel, M.; van Hellemond, J.J.; Henze, K.; Woehle, C.; Gould, S.B.; Yu, R.Y.; van der Giezen, M.; Tielens, A.G.; Martin, W.F. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 2012, 76, 444–495.
- Calvo, S.E.; Mootha, V.K. The mitochondrial proteome and human disease. Annu. Rev. Genom. Hum. Genet. 2010, 11, 25–44.
- Panigrahi, A.K.; Ogata, Y.; Zíková, A.; Anupama, A.; Dalley, R.A.; Acestor, N.; Myler, P.J.; Stuart, K.D. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 2009, 9, 434–450.
- Gawryluk, R.M.R.; Chisholm, K.A.; Pinto, D.M.; Gray, M.W. Compositional complexity of the mitochondrial proteome of a unicellular eukaryote (Acanthamoeba castellanii, supergroup Amoebozoa) rivals that of animals, fungi, and plants. J. Proteom. 2014, 109, 400–416.
- Cihlář, J.; Füssy, Z.; Horák, A.; Oborník, M. Evolution of the tetrapyrole biosynthetic pathway in secondary algae: Conservation, redundancy and replacement. PLoS ONE 2016, 11, e0166338.
- Martin, W.; Müller, M. The hydrogen hypothesis for the first eukaryote. Nature 1998, 392, 37–41.
- Boxma, B.; de Graaf, R.; van der Staay, G.; van Alen, T.A.; Ricard, G.; Gabaldón, T.; van Hoek, A.H.A.M.; van der Staay, S.Y.M.; Koopman, W.J.H.; van Hellemond, J.J.; et al. An anaerobic mitochondrion that produces hydrogen. Nature 2005, 434, 74–79.
- Martin, W.F.; Garg, S.; Zimorski, V. Endosymbiotic theories for eukaryote origin. Philos. Trans. R. Soc. B 2015, 370, 20140330.
- Cavalier-Smith, T. Origin of mitochondria by intracellular enslavement of a photosynthetic purple bacterium. Proc. Biol. Sci. 2006, 273, 1943–1952.
- Lyons, T.W.; Reinhard, C.T.; Planavsky, N.J. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 2014, 506, 307–315.
- Eme, L.; Sharpe, S.C.; Brown, M.W.; Roger, A.J. On the age of eukaryotes: Evaluating evidence from fossils and molecular clocks. Cold Spring Harb. Perspect. Biol. 2014, 6, a016139.
- Vellai, T.; Takacs, K.; Vida, G. A new aspect to the origin and evolution of eukaryotes. J. Mol. Evol. 1998, 46, 499–507.
- Kurland, C.G.; Andersson, S.G. Origin and evolution of the mitochondrial proteome. Microbiol. Mol. Biol. Rev. 2000, 64, 786–820.
- Zimorski, V.; Mentel, M.; Tielens, A.G.M.; Martin, W.F. Energy metabolism in nanerobic eukaryotes and Earth’s late oxygenation. Free Radic. Biol. Med. 2019, 140, 279–294.
- Imlay, J.A. How oxygen damages microbes: Oxygen tolerance and obligate anaerobiosis. Adv. Microb. Physiol. 2002, 46, 111–153.
- Wang, Z.; Wu, M. Phylogenomic reconstruction indicates mitochondrial ancestor was an energy parasite. PLoS ONE 2014, 9, e110685.
This entry is offline, you can click here to edit this entry!
