Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Genetics & Heredity
Hox genes play key roles in axial patterning and regulating the regional identity of cells and tissues in a wide variety of animals from invertebrates to vertebrates.
- Hox genes
- transcriptional regulation
- transcription factors
1. Introduction
Animals display remarkable variety in their body plans and there is great interest in understanding the degree to which conserved and distinct mechanisms underlie this diversity in the formation and elaboration of basic body plans in animal evolution. In chordate evolution, there is emerging evidence for a deeply conserved regulatory network, involving transcription factors (TFs) and signaling pathways, that governs patterning along the anterior–posterior (A–P) body axis [1][2][3][4][5][6]. Remarkably, despite very different morphologies among chordates, many key TFs and components of major signaling pathways (e.g., Wnts and FGFs), known to regulate developmental processes, have been shown to be similarly aligned along the A–P axis. This suggests that regulatory interactions between signaling pathways and core TFs set up a conserved gene regulatory network (GRN) that guides the formation of the basic body plan and patterning of the A–P axis. However, the question of how TFs are coupled to these ancient signaling pathways and how they integrate responses to signaling gradients is not fully understood.
The highly conserved HOX family of TFs are an example of TFs that are coupled to this ancient GRN. Hox genes are known to play key roles in axial patterning and regulating the regional identity of cells and tissues in a wide variety of animals from invertebrates to vertebrates [7][8][9][10][11]. The clustered Hox genes exhibit an interesting property known as collinearity [12][13][14][15][16]. Genes in the four mammalian Hox clusters are all transcribed in the same 5′ to 3′ direction with respect to transcription, and the order of Hox genes in each cluster on a chromosome corelates with their temporal and spatial expression domains and functions along the A–P axis of developing embryos (Figure 1). These nested domains of expression generate a combinatorial Hox code, which provides a molecular framework that serves as a key regulatory step in specifying regional identities and properties of tissues along the A–P axis. A wide variety of studies in different species and cell culture models have revealed that the nested domains of Hox expression along the A–P axis arise in part through the ability of Hox clusters to integrate and respond to opposing signaling gradients, such as those of Retinoic acid (RA), Fibroblast growth factors (Fgfs) and Wingless related integration sites (WNTs) [5][17][18][19][20][21][22][23][24][25][26][27][28][29]. Hence, it is important to understand the regulatory mechanisms through which signaling pathways are able to coordinately control the precise patterns of the transcription of the clustered Hox genes required for their roles in specifying diverse morphologic features along the A–P axis.
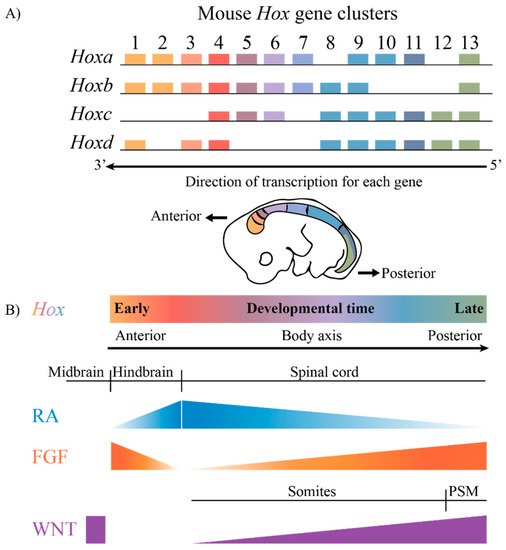
Figure 1. The mammalian Hox gene clusters and the conserved signaling pathways that play a role in defining the Hox gene expression profiles. (A) In mammals, there are four clusters of Hox genes, each on different chromosomes. They exhibit spatial and temporal collinearity, such that 3′ Hox genes are expressed early in development as well as more anteriorly in an embryo generating nested domains of expression as depicted in the drawing of an E10 mouse embryo. (B) The restricted domains of Hox expression arise through an integration of signaling molecules such as RA, FGF and WNT, which are expressed in gradients along the embryonic axis. PSM, presomitic mesoderm.
In the case of RA signaling, Hox genes are direct transcriptional targets of retinoids, and their response to RA signaling involves retinoic acid response elements (RAREs) embedded within and adjacent to the Hox clusters [18][30][31][32]. These RAREs are cis-regulatory components of RA-dependent enhancers that provide regulatory inputs both locally on adjacent Hox genes and over a long range to coordinately regulate multiple genes in a Hox cluster [33][34][35][36][37][38]. This tightly clustered organization of cis-regulatory elements and the Hox genes they control raises interesting questions with respect to roles for chromosome topology, epigenetic modifications, dynamics of transcription and the underlying transcriptional mechanisms for how enhancers display selectivity or competition between genes, and they may be shared by multiple genes in a cluster [39]. It is important that these diverse aspects of transcriptional regulation are properly coordinated to ensure the right spatial and temporal patterns and appropriate levels of expression needed for their roles in axial patterning.
The advent of new technologies for investigating the dynamics of interactions that underlie the activation of transcription are generating surprising findings. These observations challenge the widely postulated role of stable long-term enhancer promoter interactions and the notion of a single RNA polymerase with a small number of components regulating transcription [40][41][42][43][44][45][46][47][48][49]. New models suggest that dynamic condensates and mechanisms involving a series of rapid and complex interactions underlie the activation of transcription and the regulation of gene expression. It will be interesting and important to understand how this newly emerging picture of the dynamic molecular mechanisms governing transcription plays a role in modulating the inputs controlling the coordinated expression of the clustered Hox genes.
2. Regulatory Features
2.1. Enhancers
Enhancers were first discovered in simian virus 40 (SV40), where it was found that they function in an orientation-independent manner to stimulate transcription on heterologous genes [50]. Since then, a variety of analyses have revealed that animal genomes contain a large number of putative enhancers, out numbering coding genes [51][52]. It is challenging to identify cis-regulatory elements, such as enhancers, encoded in the genome through sequence analyses and computational methods alone [53]. Major efforts have been made to find ways of identifying and characterizing enhancers and their properties on a genome-wide and individual basis, which is important to facilitate our ability to decode regulatory information embedded in the genome [54][55][56][57][58][59][60]. While many development specific enhancers, including some of those discovered in the Hox clusters, are evolutionarily conserved [35][61][62][63], many adult or tissue-specific enhancers can be highly variable across species [64][65]. Even when enhancers are highly conserved, it can be challenging to understand the information content and the critical arrangements of the cis-elements that govern their ability to regulate expression [54][59][60]. Furthermore, highly conserved patterns of gene expression can arise through enhancers that display divergence [65]. Enhancers serve to stimulate transcription by integrating a variety of different regulatory inputs and binding sites for TFs to confer precise temporal, spatial and cell-type specific gene expression programs. Precise regulatory outputs from enhancers do not require that upstream factors have highly restricted domains of expression and can arise through the cumulative integration of weak, imprecise or wide-spread inputs by TFs [66][67]. The convergence of inputs can result in the integration of disparate and very broad patterns of regulatory signals into robust and tightly controlled specific outputs. Similarly, clusters of weak enhancers can synergize to serve as super enhancers to robustly regulate gene expression [68].
Enhancers can be located directly upstream of a gene or up to over a megabase away from its target gene promoter [69][70]. They frequently reside within introns of genes, even in ones they do not regulate, [70], and there is evidence for enhancers and cis-regulatory elements embedded in coding exons, including those of Hox genes [71][72][73]. Studies have shown that enhancer regions are themselves transcriptionally active. Several groups have demonstrated that non-coding enhancer RNAs are more than just transcriptional noise or byproducts of the transcriptional machinery, but are useful indicators in predicting active enhancers [74].
A challenge in identifying the targets of enhancer activity is that they can function independently of their orientation with respect to target genes and can make long-range enhancer–promoter contacts to more than the near adjacent genes [75]. Typically, an average vertebrate enhancer can be ~5 to 50 kb away from target promoters and ~1 to 10 kb away in the more compact Drosophila genome [39]. Intriguingly, the proximity of an enhancer to its target promoter required for functional activity is variable. Hence, there are no clear rules on how close enhancers should be positioned relative to the promoters they activate. Some studies have shown through proximity-dependent ligation techniques, such as 3C (Chromatin conformation capture), that enhancers physically come into contact with promoters, resulting in the activation of gene transcription [76]. Imaging approaches have shown that following the activation of genes, the distance between the enhancers and their target promoter tends to increase, suggesting a change in their interactions dependent upon their activity state [77]. This raises fundamental questions, such as, how do enhancers locate and distinguish between the target genes they activate; what confers enhancer–promoter specificity; and what degree of proximity is essential for the enhancer interactions required for gene regulation [39][55][69][75][78][79][80]?
These questions are relevant to understanding the regulation of the Hox clusters because of the high gene density and compact nature of the clusters. The enhancers embedded within and flanking an individual Hox cluster can display selective preferences, competition between genes and can regulate both near adjacent genes or act more globally on other genes in the complex. For example, in the mouse Hoxb complex, there are three RAREs in the middle of the cluster, two upstream and one downstream of Hoxb4 (Figure 2A), which participate in mediating its response to RA by regulating multiple coding and long non-coding (lncRNAs) transcripts [33][35][37][81]. One of these RAREs (DE-RARE) is an essential cis element of an RA-dependent enhancer, which undergoes epigenetic modifications, and is required to coordinate the global regulation of Hoxb genes in hematopoietic stems cells [34]. This functional role for an enhancer raises many questions regarding the mechanisms through which the DE-RARE participates in regulating so many transcripts, how targets are selected and the dynamics of the process. Why, in contrast, do other enhancers embedded in the Hoxb cluster only appear to work on a single near adjacent gene [37][62][82][83][84]?
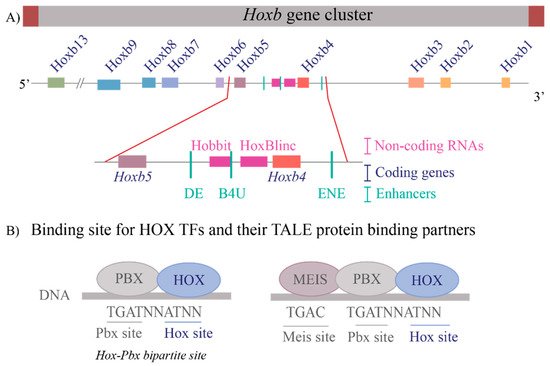
Figure 2. Transcriptional complexity of the Hoxb gene cluster and binding of HOX Transcription factors to DNA (A) A drawing of the Hoxb gene cluster to illustrate that non-coding RNAs as well as enhancers that contain RAREs (Retinoic Acid Response Elements) are interspersed within the coding Hox genes. The enlargement of the Hoxb4-Hoxb5 region shows the complexity within the region that contains three RAREs, two present upstream of Hoxb4 and one present downstream of Hoxb4 and two non-coding RNAs, Hobbit and HoxBlinc. Brown boxes flank the cluster depict boundary elements, colored squares are different Hox genes, pink boxes are non-coding RNAs, and green lines represent RARE enhancers. (B) Depicts the consensus DNA binding sites for HOX proteins and their binding partners, the TALE proteins PBX and MEIS. HOX proteins can bind on Hox-Pbx bipartite sites, or they can bind on DNA in ternary complexes along with both PBX and MEIS. Blue ovals are HOX proteins, and grey ovals are TALE protein binding partners.
This entry is adapted from the peer-reviewed paper 10.3390/jdb10010004
References
- Swalla, B.J. Building divergent body plans with similar genetic pathways. Heredity 2006, 97, 235–243.
- Lowe, C.J.; Clarke, D.N.; Medeiros, D.M.; Rokhsar, D.S.; Gerhart, J. The deuterostome context of chordate origins. Nature 2015, 520, 456–465.
- Lowe, C.J.; Wu, M.; Salic, A.; Evans, L.; Lander, E.; Stange-Thomann, N.; Gruber, C.E.; Gerhart, J.; Kirschner, M. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell 2003, 113, 853–865.
- Pani, A.M.; Mullarkey, E.E.; Aronowicz, J.; Assimacopoulos, S.; Grove, E.A.; Lowe, C.J. Ancient deuterostome origins of vertebrate brain signalling centres. Nature 2012, 483, 289–294.
- Darras, S.; Fritzenwanker, J.H.; Uhlinger, K.R.; Farrelly, E.; Pani, A.M.; Hurley, I.A.; Norris, R.P.; Osovitz, M.; Terasaki, M.; Wu, M.; et al. Anteroposterior axis patterning by early canonical Wnt signaling during hemichordate development. PLoS Biol. 2018, 16, e2003698.
- Yao, Y.; Minor, P.J.; Zhao, Y.T.; Jeong, Y.; Pani, A.M.; King, A.N.; Symmons, O.; Gan, L.; Cardoso, W.V.; Spitz, F.; et al. Cis-regulatory architecture of a brain signaling center predates the origin of chordates. Nat. Genet. 2016, 48, 575–580.
- Carroll, S.B.; Weatherbee, S.D.; Langeland, J.A. Homeotic genes and the regulation and evolution of insect wing number. Nature 1995, 375, 58–61.
- Carroll, S.B. Homeotic genes and the evolution of arthropods and chordates. Nature 1995, 376, 479–485.
- Mallo, M.; Wellik, D.M.; Deschamps, J. Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 2010, 344, 7–15.
- McGinnis, W.; Krumlauf, R. Homeobox genes and axial patterning. Cell 1992, 68, 283–302.
- Arendt, D. Hox genes and body segmentation. Science 2018, 361, 1310–1311.
- Duboule, D.; Dolle, P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989, 8, 1497–1505.
- Gaunt, S.J.; Sharpe, P.T.; Duboule, D. Spatially restricted domains of homeo-gene transcripts in mouse embryos: Relation to a segmented body plan. Development 1988, 104, 169–181.
- Graham, A.; Papalopulu, N.; Krumlauf, R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 1989, 57, 367–378.
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570.
- Dollé, P.; Izpisùa-Belmonte, J.C.; Falkenstein, H.; Renucci, A.; Duboule, D. Co-ordinate expression of the murine Hox-5 complex homeobox-containing genes during limb pattern formation. Nature 1989, 342, 767–772.
- Mallo, M.; Alonso, C.R. The regulation of Hox gene expression during animal development. Development 2013, 140, 3951–3963.
- Nolte, C.; De Kumar, B.; Krumlauf, R. Hox genes: Downstream “effectors” of retinoic acid signaling in vertebrate embryogenesis. Genesis 2019, 57, e23306.
- Parker, H.J.; Krumlauf, R. Segmental arithmetic: Summing up the Hox gene regulatory network for hindbrain development in chordates. Wiley Interdiscip. Rev. Dev. Biol. 2017, 6, e286.
- Frank, D.; Sela-Donenfeld, D. Hindbrain induction and patterning during early vertebrate development. Cell. Mol. Life Sci. 2019, 76, 941–960.
- Deschamps, J.; van Nes, J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development 2005, 132, 2931–2942.
- Deschamps, J.; Duboule, D. Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes Dev. 2017, 31, 1406–1416.
- Diez del Corral, R.; Storey, K.G. Opposing FGF and retinoid pathways: A signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. Bioessays 2004, 26, 857–869.
- Wilson, V.; Olivera-Martinez, I.; Storey, K.G. Stem cells, signals and vertebrate body axis extension. Development 2009, 136, 1591–1604.
- Young, T.; Rowland, J.E.; van de Ven, C.; Bialecka, M.; Novoa, A.; Carapuco, M.; van Nes, J.; de Graaff, W.; Duluc, I.; Freund, J.N.; et al. Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev. Cell 2009, 17, 516–526.
- Simeone, A.; Acampora, D.; Arcioni, L.; Andrews, P.W.; Boncinelli, E.; Mavilio, F. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature 1990, 346, 763–766.
- Schilling, T.F.; Nie, Q.; Lander, A.D. Dynamics and precision in retinoic acid morphogen gradients. Curr. Opin. Genet. Dev. 2012, 22, 562–569.
- Bedois, A.M.H.; Parker, H.J.; Krumlauf, R. Retinoic Acid Signaling in Vertebrate Hindbrain Segmentation: Evolution and Diversification. Diversity 2021, 13, 398.
- Krumlauf, R.; Wilkinson, D.G. Segmentation and patterning of the vertebrate hindbrain. Development 2021, 148, dev186460.
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schutz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P.; et al. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835–839.
- Rhinn, M.; Dolle, P. Retinoic acid signalling during development. Development 2012, 139, 843–858.
- Niederreither, K.; Dolle, P. Retinoic acid in development: Towards an integrated view. Nat. Rev. Genet. 2008, 9, 541–553.
- Ahn, Y.; Mullan, H.E.; Krumlauf, R. Long-range regulation by shared retinoic acid response elements modulates dynamic expression of posterior Hoxb genes in CNS development. Dev. Biol. 2014, 388, 134–144.
- Qian, P.; De Kumar, B.; He, X.C.; Nolte, C.; Gogol, M.; Ahn, Y.; Chen, S.; Li, Z.; Xu, H.; Perry, J.M.; et al. Retinoid-Sensitive Epigenetic Regulation of the Hoxb Cluster Maintains Normal Hematopoiesis and Inhibits Leukemogenesis. Cell Stem Cell 2018, 22, 740–754.e7.
- Nolte, C.; Jinks, T.; Wang, X.; Martinez Pastor, M.T.; Krumlauf, R. Shadow enhancers flanking the HoxB cluster direct dynamic Hox expression in early heart and endoderm development. Dev. Biol. 2013, 383, 158–173.
- Oosterveen, T.; Niederreither, K.; Dolle, P.; Chambon, P.; Meijlink, F.; Deschamps, J. Retinoids regulate the anterior expression boundaries of 5’ Hoxb genes in posterior hindbrain. EMBO J. 2003, 22, 262–269.
- Sharpe, J.; Nonchev, S.; Gould, A.; Whiting, J.; Krumlauf, R. Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. EMBO J. 1998, 17, 1788–1798.
- Gould, A.; Morrison, A.; Sproat, G.; White, R.A.; Krumlauf, R. Positive cross-regulation and enhancer sharing: Two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 1997, 11, 900–913.
- Furlong, E.E.M.; Levine, M. Developmental enhancers and chromosome topology. Science 2018, 361, 1341–1345.
- Narayanan, A.; Meriin, A.; Andrews, J.O.; Spille, J.H.; Sherman, M.Y.; Cisse, I.I. A first order phase transition mechanism underlies protein aggregation in mammalian cells. eLife 2019, 8, e39695.
- Henninger, J.E.; Oksuz, O.; Shrinivas, K.; Sagi, I.; LeRoy, G.; Zheng, M.M.; Andrews, J.O.; Zamudio, A.V.; Lazaris, C.; Hannett, N.M.; et al. RNA-Mediated Feedback Control of Transcriptional Condensates. Cell 2021, 184, 207–225.e24.
- Guo, Y.E.; Manteiga, J.C.; Henninger, J.E.; Sabari, B.R.; Dall’Agnese, A.; Hannett, N.M.; Spille, J.H.; Afeyan, L.K.; Zamudio, A.V.; Shrinivas, K.; et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 2019, 572, 543–548.
- Zamudio, A.V.; Dall’Agnese, A.; Henninger, J.E.; Manteiga, J.C.; Afeyan, L.K.; Hannett, N.M.; Coffey, E.L.; Li, C.H.; Oksuz, O.; Sabari, B.R.; et al. Mediator Condensates Localize Signaling Factors to Key Cell Identity Genes. Mol. Cell 2019, 76, 753–766.e6.
- Sharp, P.A.; Chakraborty, A.K.; Henninger, J.E.; Young, R.A. RNA in formation and regulation of transcriptional condensates. RNA 2021, 28, 52–57.
- Mir, M.; Stadler, M.R.; Ortiz, S.A.; Hannon, C.E.; Harrison, M.M.; Darzacq, X.; Eisen, M.B. Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. eLife 2018, 7, e40497.
- Berrocal, A.; Lammers, N.C.; Garcia, H.G.; Eisen, M.B. Kinetic sculpting of the seven stripes of the Drosophila even-skipped gene. eLife 2020, 9, e61635.
- Chong, S.; Dugast-Darzacq, C.; Liu, Z.; Dong, P.; Dailey, G.M.; Cattoglio, C.; Heckert, A.; Banala, S.; Lavis, L.; Darzacq, X.; et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 2018, 361, aar2555.
- Liu, Z.; Tjian, R. Visualizing transcription factor dynamics in living cells. J. Cell Biol. 2018, 217, 1181–1191.
- Zhang, Z.; Tjian, R. Measuring dynamics of eukaryotic transcription initiation: Challenges, insights and opportunities. Transcription 2018, 9, 159–165.
- Banerji, J.; Rusconi, S.; Schaffner, W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 1981, 27, 299–308.
- Shen, Y.; Yue, F.; McCleary, D.F.; Ye, Z.; Edsall, L.; Kuan, S.; Wagner, U.; Dixon, J.; Lee, L.; Lobanenkov, V.V.; et al. A map of the cis-regulatory sequences in the mouse genome. Nature 2012, 488, 116–120.
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74.
- Pennacchio, L.A.; Bickmore, W.; Dean, A.; Nobrega, M.A.; Bejerano, G. Enhancers: Five essential questions. Nat. Rev. Genet. 2013, 14, 288–295.
- Avsec, Z.; Weilert, M.; Shrikumar, A.; Krueger, S.; Alexandari, A.; Dalal, K.; Fropf, R.; McAnany, C.; Gagneur, J.; Kundaje, A.; et al. Base-resolution models of transcription-factor binding reveal soft motif syntax. Nat. Genet. 2021, 53, 354–366.
- Zeitlinger, J. Seven myths of how transcription factors read the cis-regulatory code. Curr. Opin. Syst. Biol. 2020, 23, 22–31.
- He, Q.; Johnston, J.; Zeitlinger, J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat. Biotechnol. 2015, 33, 395–401.
- Girskis, K.M.; Stergachis, A.B.; DeGennaro, E.M.; Doan, R.N.; Qian, X.; Johnson, M.B.; Wang, P.P.; Sejourne, G.M.; Nagy, M.A.; Pollina, E.A.; et al. Rewiring of human neurodevelopmental gene regulatory programs by human accelerated regions. Neuron 2021, 109, 3239–3251.e7.
- Snetkova, V.; Pennacchio, L.A.; Visel, A.; Dickel, D.E. Perfect and imperfect views of ultraconserved sequences. Nat. Rev. Genet. 2021.
- Snetkova, V.; Ypsilanti, A.R.; Akiyama, J.A.; Mannion, B.J.; Plajzer-Frick, I.; Novak, C.S.; Harrington, A.N.; Pham, Q.T.; Kato, M.; Zhu, Y.; et al. Ultraconserved enhancer function does not require perfect sequence conservation. Nat. Genet. 2021, 53, 521–528.
- Fuqua, T.; Jordan, J.; van Breugel, M.E.; Halavatyi, A.; Tischer, C.; Polidoro, P.; Abe, N.; Tsai, A.; Mann, R.S.; Stern, D.L.; et al. Dense and pleiotropic regulatory information in a developmental enhancer. Nature 2020, 587, 235–239.
- Marshall, H.; Studer, M.; Popperl, H.; Aparicio, S.; Kuroiwa, A.; Brenner, S.; Krumlauf, R. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature 1994, 370, 567–571.
- Parker, H.J.; De Kumar, B.; Green, S.A.; Prummel, K.D.; Hess, C.; Kaufman, C.K.; Mosimann, C.; Wiedemann, L.M.; Bronner, M.E.; Krumlauf, R. A Hox-TALE regulatory circuit for neural crest patterning is conserved across vertebrates. Nat. Commun. 2019, 10, 1189.
- Tümpel, S.; Maconochie, M.; Wiedemann, L.M.; Krumlauf, R. Conservation and diversity in the cis-regulatory networks that integrate information controlling expression of Hoxa2 in hindbrain and cranial neural crest cells in vertebrates. Dev. Biol. 2002, 246, 45–56.
- Schmidt, D.; Wilson, M.D.; Ballester, B.; Schwalie, P.C.; Brown, G.D.; Marshall, A.; Kutter, C.; Watt, S.; Martinez-Jimenez, C.P.; Mackay, S.; et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 2010, 328, 1036–1040.
- Paris, M.; Kaplan, T.; Li, X.Y.; Villalta, J.E.; Lott, S.E.; Eisen, M.B. Extensive divergence of transcription factor binding in Drosophila embryos with highly conserved gene expression. PLoS Genet. 2013, 9, e1003748.
- Farley, E.K.; Olson, K.M.; Zhang, W.; Brandt, A.J.; Rokhsar, D.S.; Levine, M.S. Suboptimization of developmental enhancers. Science 2015, 350, 325–328.
- Crocker, J.; Abe, N.; Rinaldi, L.; McGregor, A.P.; Frankel, N.; Wang, S.; Alsawadi, A.; Valenti, P.; Plaza, S.; Payre, F.; et al. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 2015, 160, 191–203.
- Choi, J.; Lysakovskaia, K.; Stik, G.; Demel, C.; Söding, J.; Tian, T.V.; Graf, T.; Cramer, P. Evidence for additive and synergistic action of mammalian enhancers during cell fate determination. eLife 2021, 10, e65381.
- Marsman, J.; Horsfield, J.A. Long distance relationships: Enhancer-promoter communication and dynamic gene transcription. Biochim. Biophys. Acta 2012, 1819, 1217–1227.
- Lettice, L.A.; Heaney, S.J.; Purdie, L.A.; Li, L.; de Beer, P.; Oostra, B.A.; Goode, D.; Elgar, G.; Hill, R.E.; de Graaff, E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003, 12, 1725–1735.
- Tümpel, S.; Cambronero, F.; Sims, C.; Krumlauf, R.; Wiedemann, L.M. A regulatory module embedded in the coding region of Hoxa2 controls expression in rhombomere 2. Proc. Natl. Acad. Sci. USA 2008, 105, 20077–20082.
- Lampe, X.; Samad, O.A.; Guiguen, A.; Matis, C.; Remacle, S.; Picard, J.J.; Rijli, F.M.; Rezsohazy, R. An ultraconserved Hox-Pbx responsive element resides in the coding sequence of Hoxa2 and is active in rhombomere 4. Nucleic Acids Res. 2008, 36, 3214–3225.
- Lin, M.F.; Kheradpour, P.; Washietl, S.; Parker, B.J.; Pedersen, J.S.; Kellis, M. Locating protein-coding sequences under selection for additional, overlapping functions in 29 mammalian genomes. Genome Res. 2011, 21, 1916–1928.
- Arnold, P.R.; Wells, A.D.; Li, X.C. Diversity and Emerging Roles of Enhancer RNA in Regulation of Gene Expression and Cell Fate. Front. Cell Dev. Biol. 2020, 7, 377.
- Schoenfelder, S.; Fraser, P. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 2019, 20, 437–455.
- Lai, F.; Gardini, A.; Zhang, A.; Shiekhattar, R. Integrator mediates the biogenesis of enhancer RNAs. Nature 2015, 525, 399–403.
- Benabdallah, N.S.; Williamson, I.; Illingworth, R.S.; Kane, L.; Boyle, S.; Sengupta, D.; Grimes, G.R.; Therizols, P.; Bickmore, W.A. Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol. Cell 2019, 76, 473–484.e7.
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637.
- Sanyal, A.; Lajoie, B.R.; Jain, G.; Dekker, J. The long-range interaction landscape of gene promoters. Nature 2012, 489, 109–113.
- Long, H.K.; Prescott, S.L.; Wysocka, J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell 2016, 167, 1170–1187.
- De Kumar, B.; Parrish, M.E.; Slaughter, B.D.; Unruh, J.R.; Gogol, M.; Seidel, C.; Paulson, A.; Li, H.; Gaudenz, K.; Peak, A.; et al. Analysis of dynamic changes in retinoid-induced transcription and epigenetic profiles of murine Hox clusters in ES cells. Genome Res. 2015, 25, 1229–1243.
- Maconochie, M.K.; Nonchev, S.; Manzanares, M.; Marshall, H.; Krumlauf, R. Differences in Krox20-dependent regulation of Hoxa2 and Hoxb2 during hindbrain development. Dev. Biol. 2001, 233, 468–481.
- Popperl, H.; Bienz, M.; Studer, M.; Chan, S.K.; Aparicio, S.; Brenner, S.; Mann, R.S.; Krumlauf, R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell 1995, 81, 1031–1042.
- Manzanares, M.; Cordes, S.; Kwan, C.-T.; Sham, M.-H.; Barsh, G.; Krumlauf, R. Segmental regulation of Hoxb3 by kreisler. Nature 1997, 387, 191–195.
This entry is offline, you can click here to edit this entry!
