The assembly of synaptic protein-DNA complexes by specialized proteins is critical for bringing together two distant sites within a DNA molecule or bridging two DNA molecules. The assembly of such synaptosomes is needed in numerous genetic processes requiring the interactions of two or more sites. The molecular mechanisms by which proteins bring the sites together, enabling the assembly of synaptosomes, remain unknown. Such proteins can utilize sliding, jumping, and segmental transfer pathways for the single-site search process, but none of these pathways explains how the synaptosome assembles. Here we used restriction enzyme SfiI, that requires the assembly of synaptosome for DNA cleavage, as our experimental system and applied time-lapse high-speed AFM (HS-AFM) to directly visualize the site search process accomplished by the SfiI enzyme.
1. Introduction
A critical step in fundamental genetic processes is the interaction between distant DNA regions. This step is controlled by specialized proteins or protein complexes which are responsible for forming site-specific protein-DNA synaptic complexes (i.e., synaptosomes)
[1]. The formation of a synaptic complex is a general phenomenon found in gene regulation (e.g., the GalR repressor)
[2], site-specific recombination (e.g., Flp, Cre recombinases)
[3], and various eukaryotic genome rearrangement systems, such as transposons, the Variable Diversity Joining (V(D)J) recombination system involving tetrameric RAG1/2 assembly
[4], and HIV integration systems
[5][6]. Notably, type II DNA restriction enzymes form synaptic complexes to complete site-specific DNA cleavage
[7]. However, little is known regarding the molecular mechanisms that underlie how such proteins search for DNA sites during the formation of synaptosomes
[1][3][7][8]. A seminal theoretical model by Berg, von Hippel, and Winter (i.e., the BHW model)
[9][10][11][12][13] describes the one-site search process using sliding, jumping, and intersegmental transfer actions as the primary pathways. However, these specific pathways cannot explain the assembly of synaptosomes, for which one protein holds two or more specific sites on the DNA substrate.
If the specific sites needed for the formation of the synaptosome are located on the same DNA molecule (i.e., pairing in
cis), the search for the two sites leads to the formation of DNA loops. The formation of such loops for various synaptosomes has been visualized with Atomic Force Microscopy (AFM) in our previous papers
[2][14][15][16][17][18][19]. Note article
[18], in which the assembly of the synaptosome with three DNA binding sites held by the EcoRII restriction enzyme was directly visualized. We also applied time-lapse high-speed AFM (HS-AFM) to visualize the dynamics of synaptic complexes assembled by EcoRII
[20]. In addition to sliding and jumping, when bound to a single site, we visualized the growth of looped complexes, in which the protein bound two separate DNA segments and searched by threading the DNA. This mechanism is specific for synaptic protein-DNA complexes, and poses the following questions: What is the contribution of this pathway to the site-search? Does the search mechanism for the synaptic system utilize a site-transfer pathway? How do they all contribute to the entire site-search process?
To address these questions, we selected the SfiI restriction enzyme as our model system. SfiI is a type IIF restriction enzyme and assembles as a homotetramer
[21] that specifically binds two sites on the DNA, each recognition site contains 13 base pairs
[22][23][24]. Our previous single molecule studies revealed the arrangement of the DNA duplexes in the SfiI synaptosome
[14], which corroborates with crystallographic data for the SfiI synaptosome
[24]. We also used HS-AFM to directly visualize concerted cleavage of DNA in the synaptosome by SfiI
[16]. Recently we characterized the efficiency of the formation of looped SfiI synaptosomes depending on the distance between the cognate sites on the DNA
[17].
2. Specific SfiI-DNA Synaptic Complexes with Different Loop Sizes
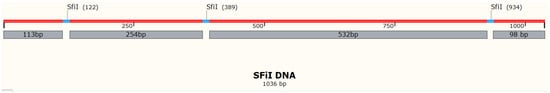
We used a 1036 bp DNA substrate containing three recognition sites for SfiI (
Figure 1) to characterize the search mechanism of SfiI. Assembly of intramolecular synaptic complexes on this substrate can lead to the formation of specific looped SfiI-DNA complexes with loop sizes of 254 bp, 532 bp, and 786 bp
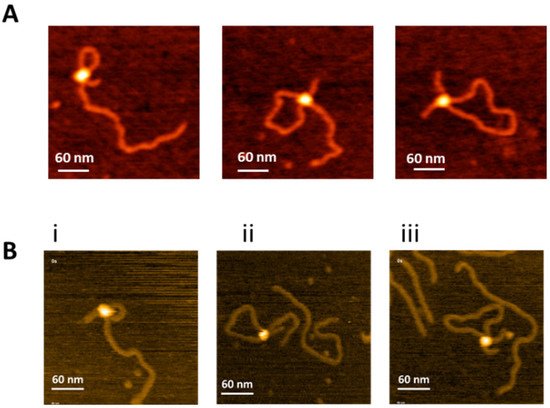
[17]. AFM images of each complex are shown in
Figure 2A. The protein appears as a bright feature connecting the two recognition sites on the DNA substrate. According to our previous studies
[17], these are highly specific synaptic complexes with high stability compared with non-specific complexes.
Figure 1. Schematic of the DNA construct with three SfiI sites. The SfiI sites are represented as blue horizontal bars. Numbers indicate size or distance (red bar) in base pair.
Figure 2. Specific SfiI-DNA synaptic complexes with different loop sizes. (
A) Left to right, dry AFM images of SfiI-DNA synaptic complexes with small, medium, and long loops sizes of 254 bp, 532 bp, and 786 bp, respectively. (
B) Initial frames of HS-AFM movies (
Movie S2a–c) showing the SfiI-DNA synaptic complexes with 254 bp (
i) 532 bp (
ii) and 786 bp (
iii) loops.
Images in
Figure 2A were captured from dry samples. However, similar specific complexes were detected in HS-AFM studies, in which images were acquired in buffer during continuous scanning of the sample. Examples of specific complexes taken from
Movies S1a–c are shown in
Figure 2B. Plate (i) and
Movie S2a correspond to the assembly of a complex with a 254 bp loop that remained stable during the observation time. Similar results were obtained for a 532 bp loop (plate ii and
Movie S2b) and a 786 bp loop (plate iii and
Movie S2c). The looped complexes formed by SfiI are the key features of synaptosomes, and assembly of a specific looped complex is the final step for the site-search process.
3. Dynamics of Synaptosomes: Threading Pathway
We have previously shown, for the EcoRII restriction enzyme
[20], that looped complexes can change loop size without protein dissociation. The main feature of such dynamics is that DNA translocates through one protein binding site, whereas another remains bound to a specific site. We termed this dynamic process as the threading pathway.
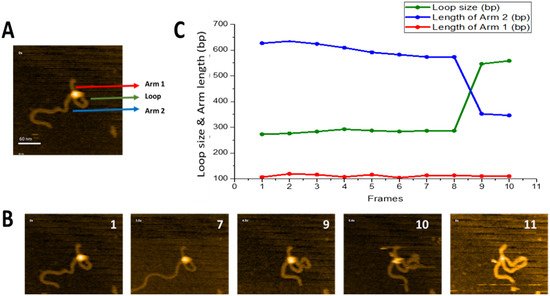
Figure 3 illustrates that SfiI is capable of this search mechanism. The full dataset is provided as
Movie S2, and a few frames are shown in
Figure 3B. A small loop is initially formed near one end of the DNA substrate (frame 1) and remains unchanged between frames 1 and 8. The loop then grows, as shown in frame 9, while the length of the short DNA flank remains unchanged. We measured the contour length of DNA and separated the measurements into the length of each flank, from the DNA ends to the protein position, and the size of the loop. The results of these measurements are presented in
Figure 3C. One flank (red line) remains stable over the observation window (112 ± 1.7 bp (SE)) and corresponds to the position of a specific site on the substrate (
Figure 1). The other flank decreases in size between frames 8 and 9 from 626 bp to 346 bp. These changes are consistent with an increase in loop size from 273 bp to 557 bp, resulting in the assembly of a specific loop. Thus, these data demonstrate that the protein, while occupying one specific site, is able to translocate via the other DNA binding site without dissociation from the specific site, which is the threading pathway for site-search.
Figure 3. Threading pathway of SfiI site-search leading to loop size increase. (
A) Initial frame of
Movie S2. SfiI tetramer is the bright feature at the cross-over of the DNA loop. The loop is highlighted with a green arrow, the small flank of the DNA (Arm 1) is highlighted with a red arrow, and the long DNA flank (Arm 2) is highlighted with a blue arrow. (
B) Progression of the loop size changes over time. The loop remains unchanged between frames 1–7, before growing in frame 9. The new larger loop remains for a single frame before SfiI dissociates in frame 11. (
C) The graph shows the change in the loop size and DNA arm lengths (in base pairs). The change in loop size and changes in the DNA Arm 1 and Arm 2 lengths are depicted in green, red, and blue, respectively.
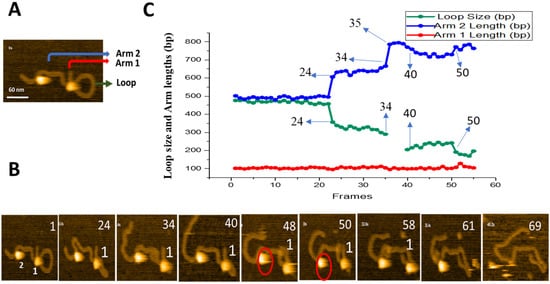
Threading pathway leading to a smaller loop was also observed, and these data are shown in
Figure 4. Initial frame with the loop size of ~450 bp is shown in
Figure 4A. A few frames from
Movie S3 are shown in
Figure 4B and the results of the contour lengths measurements for the flanks and loop sizes are plotted in
Figure 4C. The 450 bp loop remains stable for 23 frames. The loop then shrinks at frame 24, decreasing gradually between frames 24 and 34. The loop size increases at frame 40, but the loop is not stable and fluctuates in size until the protein dissociation at frame 61. The contour length measurements for different DNA segments in
Figure 4C show that the length of the short flank with 104 ± 0.7 bp (SE) remains constant over the entire observation period. The other flank changes in size, and these changes correlate with the change of the loop size. The loop size decreases from ~450 bp to 350 bp at frame 24 and continues, gradually decreasing to ~200 bp at frame 34. The loop size gradually increases between frames 40 and 50, followed by a decrease of the loop size and protein dissociation at the end of the observation period.
Figure 4. Threading pathway of SfiI site-search leading to loop size decrease. (
A) Initial frame of
movie S3. SfiI tetramer is seen as a bright feature at the center of the loop structure. The loop structure is indicated with a green arrow, the small arm of the DNA (Arm 1) is labeled with a red arrow, and the long DNA arm (Arm 2) is labeled with a blue arrow. (
B) Frames with key events. Frames 1 shows the presence of two SfiI tetramers labeled as 1 and 2. (
C) Change in DNA contour lengths in bp over time. The green line shows the change in loop size, the red line depicts the change in Arm 1 length, and the blue line shows the changes in Arm 2 length. Other pathways have been observed, and the results are described in sections below.
4. Site-Bound Segment Transfer
The HS-AFM characterization of looped SfiI-DNA synaptosomes revealed another site-search pathway, shown in
Figure 5 and
Movie S4.
Figure 5A shows the initial frame of the movie and highlights the loop and DNA flanks.
Figure 5B shows traces of the DNA to simplify the visual presentation of the images; raw images are shown in
Figure S3. The traces demonstrate that a specific synaptic complex, seen in frame 1, releases the distant recognition site between frames 2 and 3, and binds to another distant DNA segment, forming a transient loop between frames 3 and 9. SfiI then binds stably to a specific site, undergoes several transient interactions, and assembles a loop in frame 15. The loop eventually dissociates and the protein dissociates from the DNA (frame 24). We termed this pathway site-bound segmental transfer, as it resembles the intersegmental transfer pathway for the single site-search process
[8].
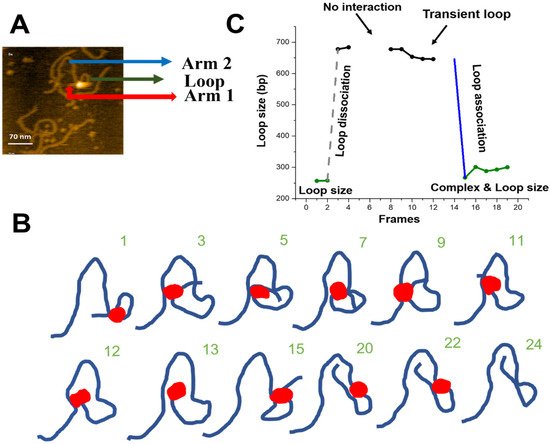
Figure 5. Site-bound segment transfer mechanism of SfiI. (
A) Initial frame of
Movie S4. The sfiI tetramer is the bright feature at the center of the loop structure. The loop structure is indicated with a green arrow, the small DNA arm (Arm 1) is shown by a red arrow, and the long DNA arm (Arm 2) is labeled with a blue arrow. (
B) Traces of the synaptic complex from selected HS-AFM movie frames. Raw images are shown in
Figure S3. (
C) Loop size change in bp ± SE versus frame number. The green indicates the loop size of the synaptic complex. Grey indicates the dissociation of the loop complex. Black indicates transient loops.
We measured the sizes of the loops and the length of the DNA flanks for the site-bound segmental transfer process, shown in Figure 5C. Arm 1 of the DNA (red arrow) remains stable (108 ± 0.9 bp (SE)) over the observation period and corresponds to the position of the specific site on the substrate (scheme in Figure 1). The initial loop size (green line) corresponds to the 256 bp specific synaptic loop. After the loop dissociation (dashed grey line), a transient ~680 bp loop is formed without protein dissociation from the specific site at 108 bp. Between frames 8 and 12, the loop shrinks slightly, from 677 bp to 645 bp. Finally, another site-bound transfer occurs (frame 15), leading to a loop close to the initial size (266 bp). Taken together, these observations indicate that SfiI uses a site-bound segment transfer mechanism to search the DNA substrate while being bound to one recognition site.
5. Sliding and Jumping Pathways
We also characterized SfiI dynamics after the loop dissociation. One such event is shown in
Figure 6, with the complete dataset assembled as
Movie S5. A few frames are shown in
Figure 6B. The initial looped complex in frame 1 dissociates after 12 consecutive frames, following which the protein translocates along the DNA duplex. As a result, by the end of the observation (frame 39), the protein translocates from the point of loop dissociation as a tetramer to a distant site on the same DNA molecule and dissociates from the DNA.
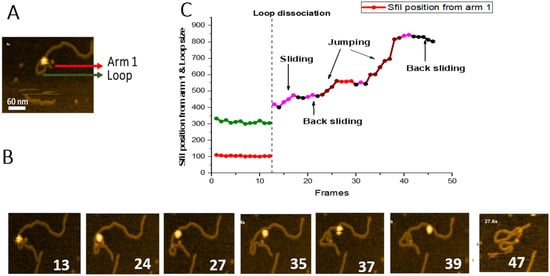
Figure 6. Sliding and jumping of SfiI tetramer after loop dissociation. (
A) Initial frame of the
movie S5. SfiI tetramer can be seen as a bright feature at the center of the loop structure. The loop is indicated with a green arrow, the small DNA arm (Arm 1) is labeled with a red arrow. (
B) HS-AFM movie frames with important events between frames 1 and 46. The frames show dissociation of the complex and translocation of the SfiI tetramer along the DNA. (
C) Graph showing the position of the SfiI tetramer along the length of the DNA measured in base pairs (bp) ± SE. The blue line indicates the length of the DNA Arm 1. The vertical dotted line indicates loop dissociation. The different events observed during were indicated with different colors, as shown in the graph.
To characterize the translocation process, we measured the SfiI position, based on the contour lengths of the DNA flanks, and plotted these data in Figure 6C. The short flank (red line), which corresponds to the location of a recognition site, remains stable (104 ± 0.83 bp (SE) until frame 12. Similarly, the loop remains relatively unchanged during this period (green line, 312 ± 2.8 bp (SE), before dissociating at frame 13. Following loop dissociation, the protein remains bound to the DNA duplex. It translocates along the duplex utilizing sliding and jumping pathways, indicated in Figure 6C, with different colors. The graph also shows that the protein slides over the DNA duplex, covering the distance between ~418 bp and 501 bp. After frame 23, SfiI jumps from position 501 bp to 561 bp. SfiI then slides back between frames 28 and 32, reaching the position at 543 bp. Another change in the protein position results from jumps between frames 33 to 39, measuring a distance of almost 300 bp (from 543 bp to 823 bp). Sliding between frames 40 and 41 results in the change of protein position from 823 bp to 842 bp. Finally, SfiI slides back between frames 42 to 45 (from 842 bp to 802 bp) and dissociates from the DNA at frame 46.
Similar pathways of SfiI site-search dynamics are shown in
Movie S6 and illustrated with key frames in
Figure S4. The SfiI position from the far end of the DNA was measured, and the graph,
Figure S4C, shows the sliding of the SfiI to the end of the DNA flank. An interesting phenomenon was also observed, in which SfiI, after dissociation from one DNA flank, jumps and binds to another end of the DNA between frames 25 and 36 before dissociating from the DNA.
Taken together our results shows that SfiI utilizes threading and site-bound segment transfer pathways to assemble the synaptosome. We have observed the sliding and jumping of SfiI, which the protein utilizes to initiate the site-search process. During the site-search, transient loops are formed, facilitating the search of another specific site. The contribution of the threading pathway in the site-search process is slightly greater than the site-bound segment transfer pathway in our observation; however, a combination of both site-bound segment transfer and threading facilitates the assembly of the protein-DNA synaptic complex.
This entry is adapted from the peer-reviewed paper 10.3390/ijms23010212