Currently it is difficult to predict the outcome of spinal cord injuries and neurodegenerative diseases diagnosed in the early stages. These difficulties can be solved with the help of biomarkers - molecules that play an important role in these pathological processes. Small non-coding RNAs, such as microRNAs have the potential to act as biomarkers. Micro-RNAs are short RNAs of ~ 22 nucleotides, which are not translated into proteins, but instead act directly to regulate the translation of protein coding messenger RNAs. Isolation of microRNAs from biological fluids, in particular CSF, is problematic. As such, our work focuses on the methodological aspects as well as reviewing both the animal and patient studies used to determine potential miRNA biomarkers. In addition, we propose possible solutions for the existing technical problems.
- neurological disorders
- spinal cord injury
- neurodegenerative diseases
- miRNA
1. Introduction
2. Methods for Isolation of miRNAs from Cerebrospinal Fluid
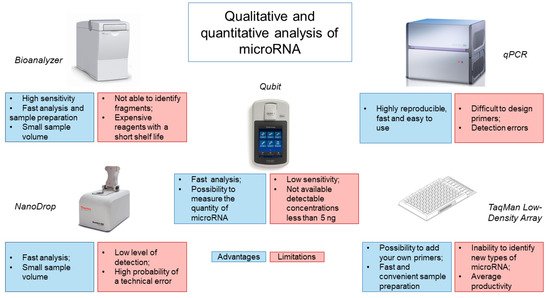
3. Assessment of the Quality of the Obtained miRNA
4. Methods for Studying the miRNA Expression Profile
This entry is adapted from the peer-reviewed paper 10.3390/ijms23010114
References
- Rao, P.; Benito, E.; Fischer, A. MicroRNAs as biomarkers for CNS disease. Front. Mol. Neurosci. 2013, 6, 39.
- Martinez, B.; Peplow, P.V. MicroRNAs as diagnostic and therapeutic tools for Alzheimer’s disease: Advances and limitations. Neural Regen. Res. 2019, 14, 242–255.
- Batistela, M.S.; Josviak, N.D.; Sulzbach, C.D.; de Souza, R.L.R. An overview of circulating cell-free microRNAs as putative biomarkers in Alzheimer’s and Parkinson’s Diseases. Int. J. Neurosci. 2017, 127, 547–558.
- Singh, A.; Tetreault, L.; Kalsi-Ryan, S.; Nouri, A.; Fehlings, M.G. Global prevalence and incidence of traumatic spinal cord injury. Clin. Epidemiol. 2014, 6, 309.
- Liu, W.Z.; Ma, Z.J.; Li, J.R.; Kang, X.W. Mesenchymal stem cell-derived exosomes: Therapeutic opportunities and challenges for spinal cord injury. Stem Cell Res. Ther. 2021, 12, 102.
- Noonan, V.K.; Fingas, M.; Farry, A.; Baxter, D.; Singh, A.; Fehlings, M.G.; Dvorak, M.F. Incidence and Prevalence of Spinal Cord Injury in Canada: A National Perspective. Neuroepidemiology 2012, 38, 219–226.
- Curt, A.; Van Hedel, H.J.A.; Klaus, D.; Dietz, V.; Grp, E.-S.S. Recovery from a spinal cord injury: Significance of compensation, neural plasticity, and repair. J. Neurotrauma 2008, 25, 677–685.
- Steeves, J.D.; Kramer, J.K.; Fawcett, J.W.; Cragg, J.; Lammertse, D.P.; Blight, A.R.; Marino, R.J.; Ditunno, J.F.; Coleman, W.P.; Geisler, F.H.; et al. Extent of spontaneous motor recovery after traumatic cervical sensorimotor complete spinal cord injury. Spinal Cord 2011, 49, 257–265.
- Sandwell, S.; Markandaya, M. Neurotrauma, Prognosis and Outcome Predictions BT—Encyclopedia of Trauma Care; Papadakos, P.J., Gestring, M.L., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1079–1082.
- Kwon, B.K.; Streijger, F.; Fallah, N.; Noonan, V.K.; Belanger, L.M.; Ritchie, L.; Paquette, S.J.; Ailon, T.; Boyd, M.C.; Street, J.; et al. Cerebrospinal Fluid Biomarkers To Stratify Injury Severity and Predict Outcome in Human Traumatic Spinal Cord Injury. J. Neurotrauma 2017, 34, 567–580.
- Ogurcov, S.; Shulman, I.; Garanina, E.; Sabirov, D.; Baichurina, I.; Kuznetcov, M.; Masgutova, G.; Kostennikov, A.; Rizvanov, A.; James, V.; et al. Blood Serum Cytokines in Patients with Subacute Spinal Cord Injury: A Pilot Study to Search for Biomarkers of Injury Severity. Brain Sci. 2021, 11, 322.
- Yunta, M.; Nieto-Diaz, M.; Esteban, F.J.; Caballero-Lopez, M.; Navarro-Ruiz, R.; Reigada, D.; Pita-Thomas, D.W.; del Aguila, A.; Munoz-Galdeano, T.; Maza, R.M. MicroRNA Dysregulation in the Spinal Cord following Traumatic Injury. PLoS ONE 2012, 7, e34534.
- Morganti, S.; Tarantino, P.; Ferraro, E.; D’Amico, P.; Viale, G.; Trapani, D.; Duso, B.A.; Curigliano, G. Complexity of genome sequencing and reporting: Next generation sequencing (NGS) technologies and implementation of precision medicine in real life. Crit. Rev. Oncol. Hematol. 2019, 133, 171–182.
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105.
- Pinchi, E.; Frati, A.; Cantatore, S.; D’Errico, S.; La Russa, R.; Maiese, A.; Palmieri, M.; Pesce, A.; Viola, R.V.; Frati, P.; et al. Acute Spinal Cord Injury: A Systematic Review Investigating miRNA Families Involved. Int. J. Mol. Sci. 2019, 20, 1841.
- Katoh, M. Cardio-miRNAs and onco-miRNAs: Circulating miRNA-based diagnostics for non-cancerous and cancerous diseases. Front. Cell Dev. Biol. 2014, 2, 61.
- Liu, N.K.; Wang, X.F.; Lu, Q.B.; Xu, X.M. Altered microRNA expression following traumatic spinal cord injury. Exp. Neurol. 2009, 219, 424–429.
- Nieto-Diaz, M.; Esteban, F.J.; Reigada, D.; Munoz-Galdeano, T.; Yunta, M.; Caballero-Loopez, M.; Navarro-Ruiz, R.; del Aguila, A.; Maza, R.M. MicroRNA dysregulation in spinal cord injury: Causes, consequences, and therapeutics. Front. Cell. Neurosci. 2014, 8, 53.
- Burgos, K.L.; Javaherian, A.; Bomprezzi, R.; Ghaffari, L.; Rhodes, S.; Courtright, A.; Tembe, W.; Kim, S.; Metpally, R.; Van Keuren-Jensen, K. Identification of extracellular miRNA in human cerebrospinal fluid by next-generation sequencing. RNA 2013, 19, 712–722.
- McAlexander, M.A.; Phillips, M.J.; Witwer, K.W. Comparison of methods for miRNA extraction from plasma and quantitative recovery of RNA from cerebrospinal fluid. Front. Genet. 2013, 4, 83.
- Bache, S.; Rasmussen, R.; Rossing, M.; Laigaard, F.P.; Nielsen, F.C.; Moller, K. MicroRNA Changes in Cerebrospinal Fluid After Subarachnoid Hemorrhage. Stroke 2017, 48, 2391.
- Akers, J.C.; Hua, W.; Li, H.Y.; Ramakrishnan, V.; Yang, Z.X.; Quan, K.; Zhu, W.; Li, J.; Figueroa, J.; Hirshman, B.R.; et al. A cerebrospinal fluid microRNA signature as biomarker for glioblastoma. Oncotarget 2017, 8, 68769–68779.
- Kopkova, A.; Sana, J.; Fadrus, P.; Slaby, O. Cerebrospinal fluid microRNAs as diagnostic biomarkers in brain tumors. Clin. Chem. Lab. Med. 2018, 56, 869–879.
- Gui, Y.X.; Liu, H.; Zhang, L.S.; Lv, W.; Hu, X.Y. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015, 6, 37043–37053.
- Saugstad, J.A.; Lusardi, T.A.; Van Keuren-Jensen, K.R.; Phillips, J.I.; Lind, B.; Harrington, C.A.; McFarland, T.J.; Courtright, A.L.; Reiman, R.A.; Yeri, A.S.; et al. Analysis of extracellular RNA in cerebrospinal fluid. J. Extracell. Vesicles 2017, 6, 1317577.
- Sorensen, S.S.; Nygaard, A.B.; Carlsen, A.L.; Heegaard, N.H.H.; Bak, M.; Christensen, T. Elevation of brain-enriched miRNAs in cerebrospinal fluid of patients with acute ischemic stroke. Transl. Stroke Res. 2017, 5, 24.
- Sorensen, S.S.; Nygaard, A.B.; Nielsen, M.Y.; Jensen, K.; Christensen, T. miRNA Expression Profiles in Cerebrospinal Fluid and Blood of Patients with Acute Ischemic Stroke. Transl. Stroke Res. 2014, 5, 711–718.
- Masotti, A.; Preckel, T. Analysis of small RNAs with the Agilent 2100 Bioanalyzer. Nat. Methods 2006, 3, 658.
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369.
- Denk, J.; Boelmans, K.; Siegismund, C.; Lassner, D.; Arlt, S.; Jahn, H. MicroRNA profiling of CSF reveals potential biomarkers to detect Alzheimers disease. PLoS ONE 2015, 10, e0126423.
- Van Harten, A.C.; Mulders, J.; Scheltens, P.; van der Flier, W.M.; Oudejans, C.B.M. Differential Expression of microRNA in Cerebrospinal Fluid as a Potential Novel Biomarker for Alzheimer’s Disease. J. Alzheimers Dis. 2015, 47, 243–252.
- Kiko, T.; Nakagawa, K.; Tsuduki, T.; Furukawa, K.; Arai, H.; Miyazawa, T. MicroRNAs in Plasma and Cerebrospinal Fluid as Potential Markers for Alzheimer’s Disease. J. Alzheimer’s Dis. 2014, 39, 253–259.
- Kodani, M.; Yang, G.Y.; Conklin, L.M.; Travis, T.C.; Whitney, C.G.; Anderson, L.J.; Schrag, S.J.; Taylor, T.H.; Beall, B.W.; Breiman, R.F.; et al. Application of TaqMan Low-Density Arrays for Simultaneous Detection of Multiple Respiratory Pathogens. J. Clin. Microbiol. 2011, 49, 2175–2182.
- Kopkova, A.; Sana, J.; Fadrus, P.; Machackova, T.; Vecera, M.; Vybihal, V.; Juracek, J.; Vychytilova-Faltejskova, P.; Smrcka, M.; Slaby, O. MicroRNA isolation and quantification in cerebrospinal fluid: A comparative methodical study. PLoS ONE 2018, 13, e0208580.
- Shalaby, T.; Grotzer, M.A. Tumor-Associated CSF MicroRNAs for the Prediction and Evaluation of CNS Malignancies. Int. J. Mol. Sci. 2015, 16, 29103–29119.
- Dangla-Valls, A.; Molinuevo, J.L.; Altirriba, J.; Sanchez-Valle, R.; Alcolea, D.; Fortea, J.; Rami, L.; Balasa, M.; Munoz-Garcia, C.; Ezquerra, M.; et al. CSF microRNA Profiling in Alzheimer’s Disease: A Screening and Validation Study. Mol. Neurobiol. 2017, 54, 6647–6654.
- Baraniskin, A.; Kuhnhenn, J.; Schlegel, U.; Maghnouj, A.; Zollner, H.; Schmiegel, W.; Hahn, S.; Schroers, R. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro Oncol. 2012, 14, 29–33.
- Baraniskin, A.; Chomiak, M.; Ahle, G.; Gress, T.; Buchholz, M.; Turewicz, M.; Eisenacher, M.; Margold, M.; Schlegel, U.; Schmiegel, W.; et al. MicroRNA-30c as a novel diagnostic biomarker for primary and secondary B-cell lymphoma of the CNS. J. Neurooncol. 2018, 137, 463–468.
- Riancho, J.; Vazquez-Higuera, J.L.; Pozueta, A.; Lage, C.; Kazimierczak, M.; Bravo, M.; Calero, M.; Gonzalez, A.; Rodriguez, E.; Lleo, A.; et al. MicroRNA Profile in Patients with Alzheimer’s Disease: Analysis of miR-9-5p and miR-598 in Raw and Exosome Enriched Cerebrospinal Fluid Samples. J. Alzheimer’s Dis. 2017, 57, 483–491.
- Yagi, Y.; Ohkubo, T.; Kawaji, H.; Machida, A.; Miyata, H.; Goda, S.; Roy, S.; Hayashizaki, Y.; Suzuki, H.; Yokota, T. Next-generation sequencing-based small RNA profiling of cerebrospinal fluid exosomes. Neurosci. Lett. 2017, 636, 48–57.
- Wang, W.X.; Fardo, D.W.; Jicha, G.A.; Nelson, P.T. A Customized Quantitative PCR MicroRNA Panel Provides a Technically Robust Context for Studying Neurodegenerative Disease Biomarkers and Indicates a High Correlation Between Cerebrospinal Fluid and Choroid Plexus MicroRNA Expression. Mol. Neurobiol. 2017, 54, 8191–8202.
- Tsang, H.F.; Xue, V.W.; Koh, S.P.; Chiu, Y.M.; Ng, L.P.W.; Wong, S.C.C. NanoString, a novel digital color-coded barcode technology: Current and future applications in molecular diagnostics. Expert Rev. Mol. Diagn. 2017, 17, 95–103.
- Wang, H.; Horbinski, C.; Wu, H.; Liu, Y.X.; Sheng, S.Y.; Liu, J.P.; Weiss, H.; Stromberg, A.J.; Wang, C. NanoStringDiff: A novel statistical method for differential expression analysis based on NanoString nCounter data. Nucleic Acids Res. 2016, 44, e151.
- Hong, L.Z.; Zhou, L.; Zou, R.; Khoo, C.M.; San Chew, A.L.; Chin, C.L.; Shih, S.J. Systematic evaluation of multiple qPCR platforms, NanoString and miRNA-Seq for microRNA biomarker discovery in human biofluids. Sci. Rep. 2021, 11, 4435.
- Tigchelaar, S.; Gupta, R.; Shannon, C.P.; Streijger, F.; Sinha, S.; Flibotte, S.; Rizzuto, M.A.; Street, J.; Paquette, S.; Ailon, T.; et al. MicroRNA Biomarkers in Cerebrospinal Fluid and Serum Reflect Injury Severity in Human Acute Traumatic Spinal Cord Injury. J. Neurotrauma 2019, 36, 2358–2371.
