Arbuscular mycorrhizal fungi (AMF) are obligate biotrophs that supply mineral nutrients to the host plant in exchange for carbon derived from photosynthesis. Sucrose is the end-product of photosynthesis and the main compound used by plants to translocate photosynthates to non-photosynthetic tissues. AMF alter carbon distribution in plants by modifying the expression and activity of key enzymes of sucrose biosynthesis, transport, and/or catabolism.
- arbuscular mycorrhiza
- mycorrhizal plants
1. Introduction
Arbuscular mycorrhiza (AM) is a mutualistic association between fungi from the Glomeromycotina group and plants from most phylogenetic clades [1][2]. The mutualistic nature of arbuscular mycorrhizae implies a bidirectional flow of nutrients between plant roots and arbuscular mycorrhizal fungi (AMF). The plant gives up part of its photoassimilates to the fungus, an obligate symbiont with a heterotrophic metabolism that grows and develops in the internal root tissues [3]. One key benefit for the host plants from this biological interaction is the improved acquisition of water and mineral nutrients, in particular phosphorus (P) [4][5][6]. The regulation of this resource exchange between the plant and fungal partners is key for the functioning of symbiosis, as it determines the net outcome of the interaction. Major research efforts have been devoted in the last few decades to an understanding of the trading of resources between the plant and fungal symbiont [7][8].
2. Sucrose Transporters and Sucrose Mobilization in Mycorrhizal Plants
3. Mycorrhizal Symbiosis and Sucrose Catabolism in Plants
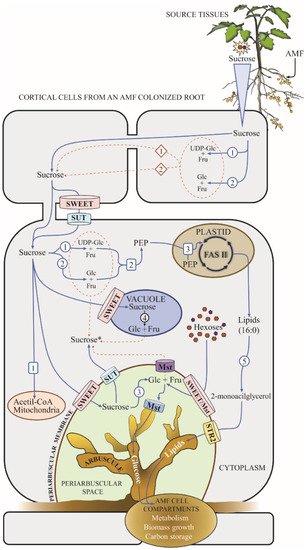
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms10010075
References
- Bonfante, P.; Genre, A. Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat. Commun. 2010, 1, 48.
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A.; et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 2016, 108, 1028–1046.
- Genre, A.; Lanfranco, L.; Perotto, S.; Bonfante, P. Unique and common traits in mycorrhizal symbioses. Nat. Rev. Microbiol. 2020, 18, 649–660.
- Smith, S.E.; Smith, F.A. Roles of arbuscular mycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 2011, 62, 227–250.
- Ferrol, N.; Azcón-Aguilar, C.; Pérez-Tienda, J. Review: Arbuscular mycorrhizas as key players in sustainable plant phosphorus acquisition: An overview on the mechanisms involved. Plant Sci. 2019, 280, 441–447.
- van Creij, J.; Wang, P.; Limpens, E. Arbuscular mycorrhiza, a fungal perspective. In Molecular aspects of plant beneficial microbes in agriculture, 1st ed.; Sharma, V., Salawan, R., Tawfeeq Al-Ani, L.K., Eds.; Elsevier: London, UK, 2020; ISBN 9780128184691.
- Konečný, J.; Hršelová, H.; Bukovská, P.; Hujslová, M.; Jansa, J. Correlative evidence for co-regulation of phosphorus and carbon exchanges with symbiotic fungus in the arbuscular mycorrhizal Medicago truncatula. PLoS ONE 2019, 14, e0224938.
- Wipf, D.; Krajinski, F.; van Tuinen, D.; Recorbet, G.; Courty, P.-E. Trading on the arbuscular mycorrhiza market: From arbuscules to common mycorrhizal networks. New Phytol. 2019, 223, 1127–1142.
- Kühn, C.; Grof, C.P.L. Sucrose transporters of higher plants. Curr. Opin. Plant Biol. 2010, 13, 287–297.
- Reinders, A.; Sivitz, A.B.; Ward, J.M. Evolution of plant sucrose uptake transporters. Front. Plant Sci. 2012, 3, 22.
- Ge, L.; Sun, S.; Chen, A.; Kapulnik, Y.; Xu, G. Tomato sugar transporter genes associated with mycorrhiza and phosphate. Plant Growth Regul. 2008, 55, 115–123.
- Boldt, K.; Pörs, Y.; Haupt, B.; Bitterlich, M.; Kühn, C.; Grimm, B.; Franken, P. Photochemical processes, carbon assimilation and RNA accumulation of sucrose transporter genes in tomato arbuscular mycorrhiza. J. Plant Physiol. 2011, 168, 1256–1263.
- Doidy, J.; Van Tuinen, D.; Lamotte, O.; Corneillat, M.; Alcaraz, G.; Wipf, D. The Medicago truncatula sucrose transporter family: Characterization and implication of key members in carbon partitioning towards arbuscular mycorrhizal fungi. Mol. Plant 2012, 5, 1346–1358.
- Gabriel-Neumann, E.; Neumann, G.; Leggewie, G.; George, E. Constitutive overexpression of the sucrose transporter SoSUT1 in potato plants increases arbuscular mycorrhiza fungal root colonization under high, but not under low, soil phosphorus availability. J. Plant Physiol. 2011, 168, 911–919.
- Bitterlich, M.; Krügel, U.; Boldt-Burisch, K.; Franken, P.; Kühn, C. The sucrose transporter SlSUT2 from tomato interacts with brassinosteroid functioning and affects arbuscular mycorrhiza formation. Plant J. 2014, 78, 877–889.
- Hansch, F.; Jaspar, H.; von Sivers, L.; Bitterlich, M.; Franken, P.; Kühn, C. Brassinosteroids and sucrose transport in mycorrhizal tomato plants. Plant Signal. Behav. 2020, 15, 1714292.
- Braun, D.M. SWEET! The pathway is complete. Science 2012, 335, 173–174.
- Feng, C.-Y.; Han, J.-X.; Han, X.-X.; Jiang, J. Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 2015, 573, 261–272.
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for pholem transport. Sciencie 2012, 335, 207–211.
- Chen, L.-Q.; Lin, I.W.; Qu, X.-Q.; Sosso, D.; McFarlane, H.E.; Londoño, A.; Samuels, A.L.; Frommer, W.B. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 2015, 27, 607–619.
- Patil, G.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genom. 2015, 16, 520.
- Iftikhar, J.; Lyu, M.; Liu, Z.; Mehmood, N.; Munir, N.; Ahmed, M.A.A.; Batool, W.; Aslam, M.M.; Yuan, Y.; Wu, B. Sugar and hormone dynamics and the expression profiles of SUT/SUC and SWEET sugar transporters during flower development in Petunia axillaris. Plants 2020, 9, 1770.
- Manck-Götzenberger, J.; Requena, N. Arbuscular mycorrhiza symbiosis induces a major transcriptional reprogramming of the potato SWEET sugar transporter family. Front. Plant Sci. 2016, 7, 487.
- Chong, J.; Piron, M.-C.; Meyer, S.; Merdinoglu, D.; Bertsch, C.; Mestre, P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J. Exp. Bot. 2014, 65, 6589–6601.
- Sugiyama, A.; Saida, Y.; Yoshimizu, M.; Takanashi, K.; Sosso, D.; Frommer, W.B.; Yazaki, K. Molecular characterization of LjSWEET3, a sugar transporter in nodules of Lotus japonicus. Plant Cell Physiol. 2017, 58, 298–306.
- Bezrutczyk, M.; Yang, J.; Eom, J.-S.; Prior, M.; Sosso, D.; Hartwig, T.; Szurek, B.; Oliva, R.; Vera-Cruz, C.; White, F.F.; et al. Sugar flux and signaling in plant-microbe interactions. Plant J. 2018, 93, 675–685.
- Kafle, A.; Garcia, K.; Wang, X.; Pfeffer, P.E.; Strahan, G.D.; Bücking, H. Nutrient demand and fungal access to resources control the carbon allocation to the symbiotic partners in tripartite interactions of Medicago truncatula. Plant Cell Environ. 2019, 42, 270–284.
- An, J.; Zeng, T.; Ji, C.; de Graaf, S.; Zheng, Z.; Xiao, T.T.; Deng, X.; Xiao, S.; Bisseling, T.; Limpens, E.; et al. A Medicago truncatula SWEET transporter implicated in arbuscule maintenance during arbuscular mycorrhizal symbiosis. New Phytol. 2019, 224, 396–408.
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532.
- Radhakrishnan, G.V.; Keller, J.; Rich, M.K.; Vernié, T.; Mbadinga Mbadinga, D.L.; Vigneron, N.; Cottret, L.; Clemente, H.S.; Libourel, C.; Cheema, J.; et al. An ancestral signalling pathway is conserved in intracellular symbioses-forming plant lineages. Nat. Plants 2020, 6, 280–289.
- Harrison, M.J. A sugar transporter from Medicago truncatula: Altered expression pattern in roots during vesicular-arbuscular (VA) mycorrhizal associations. Plant J. 1996, 9, 491–503.
- García-Rodríguez, S.; Pozo, M.J.; Azcón-Aguilar, C.; Ferrol, N. Expression of a tomato sugar transporter is increased in leaves of mycorrhizal or Phytophthora parasitica-infected plants. Mycorrhiza 2005, 15, 489–496.
- Reuscher, S.; Akiyama, M.; Yasuda, T.; Makino, H.; Aoki, K.; Shibata, D.; Shiratake, K. The sugar transporter inventory of tomato: Genome-wide identification and expression analysis. Plant Cell Physiol. 2014, 55, 1123–1141.
- Zhao, S.; Chen, A.; Chen, C.; Li, C.; Xia, R.; Wang, X. Transcriptomic analysis reveals the possible roles of sugar metabolism and export for positive mycorrhizal growth responses in soybean. Physiol. Plant. 2019, 166, 712–728.
- Wright, D.P.; Scholes, J.D.; Read, D.J.; Rolfe, S.A. European and african maize cultivars differ in their physiological and molecular responses to mycorrhizal infection. New Phytol. 2005, 167, 881–896.
- Xu, X.; Yang, Y.; Liu, C.; Sun, Y.; Zhang, T.; Hou, M.; Huang, S.; Yuan, H. The evolutionary history of the sucrose synthase gene family in higher plants. BMC Plant Biol. 2019, 19, 566.
- Tauzin, A.S.; Giardina, T. Sucrose and invertases, a part of the plant defense response to the biotic stresses. Front. Plant Sci. 2014, 5, 293.
- Schaarschmidt, S.; Roitsch, T.; Hause, B. Arbuscular mycorrhiza induces gene expression of the apoplastic invertase LIN6 in tomato (Lycopersicon esculentum) roots. J. Exp. Bot. 2006, 57, 4015–4023.
- Proels, R.K.; Roitsch, T. Extracellular invertase LIN6 of tomato: A pivotal enzyme for integration of metabolic, hormonal, and stress signals is regulated by a diurnal rhythm. J. Exp. Bot. 2009, 60, 1555–1567.
- García-Rodríguez, S.; Azcón-Aguilar, C.; Ferrol, N. Transcriptional regulation of host enzymes involved in the cleavage of sucrose during arbuscular mycorrhizal symbiosis. Physiol. Plant. 2007, 129, 737–746.
- Blee, K.A.; Anderson, A.J. Transcripts for genes encoding soluble acid invertase and sucrose synthase accumulate in root tip and cortical cells containing mycorrhizal arbuscules. Plant Mol. Biol. 2002, 50, 197–211.
- Schubert, A.; Allara, P.; Morte, A. Cleavage of sucrose in roots of soybean (Glycine max) colonized by an arbuscular mycorrhizal fungus. New Phytol. 2004, 161, 495–501.
- Roth, R.; Paszkowski, U. Plant carbon nourishment of arbuscular mycorrhizal fungi. Curr. Opin. Plant Biol. 2017, 39, 50–56.
- Schaarschmidt, S.; Kopka, J.; Ludwig-Müller, J.; Hause, B. Regulation of arbuscular mycorrhization by apoplastic invertases: Enhanced invertase activity in the leaf apoplast affects the symbiotic interaction. Plant J. 2007, 51, 390–405.
- Schaarschmidt, S.; González, M.-C.; Roitsch, T.; Strack, D.; Sonnewald, U.; Hause, B. Regulation of arbuscular mycorrhization by carbon. The symbiotic interaction cannot be improved by increased carbon availability accomplished by root-specifically enhanced invertase activity. Plant Physiol. 2007, 143, 1827–1840.
- Hohnjec, N.; Perlick, A.M.; Pühler, A.; Küster, H. The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol. Plant-Microbe Interact. 2003, 16, 903–915.
- Baier, M.C.; Keck, M.; Gödde, V.; Niehaus, K.; Küster, H.; Hohnjec, N. Knockdown of the symbiotic sucrose synthase MtSucS1 affects arbuscule maturation and maintenance in mycorrhizal roots of Medicago truncatula. Plant Physiol. 2010, 152, 1000–1014.
