DYRK (dual-specificity tyrosine-regulated kinases) are an evolutionary conserved family of protein kinases with members from yeast to humans. In humans, DYRKs are pleiotropic factors that phosphorylate a broad set of proteins involved in many different cellular processes, including those associated with all the hallmarks of cancer. In accordance with an involvement of DYRK kinases in the regulation of tumorigenic processes, an increasing number of research studies are showing either alterations of DYRK gene expression in tumor samples and/or providing evidence of DYRK-dependent mechanisms that contribute to tumor initiation and/or progression.
- DYRK kinases
- cellular signaling
- expression dysregulation
- cell cycle
- cell survival
- tumor progression
- kinase inhibitors
1. The DYRK Family of Kinases
The first cancer gene identified, the proto-oncogene c-Src, was found to encode a protein kinase [1]. Yet, since then, almost a hundred kinase genes have been attributed a tumor suppressor or oncogenic role, and they represent the most abundant class of cancer driver genes known to date [2]. Dual-specificity tyrosine-regulated kinases (DYRKs) belong to the CMGC group of kinases, which includes cyclin-dependent kinases (CDKs), mitogen-activated protein kinases (MAPKs), CDK-like kinases, the serine-arginine-rich protein kinase, Cdc2-like kinases (CLKs) and members of the RCK family [3]. The DYRK family is formed by three subfamilies: the DYRK subfamily, the homeodomain-interacting kinases (HIPKs), and the pre-messenger RNA-processing protein 4 kinases (PRP4Ks) [3]. Here, we will use “DYRK” to refer specifically to the DYRK subfamily, which contains five members in humans that are clustered into two classes based on their phylogenetic relationships [4]: class I DYRKs, DYRK1A and DYRK1B (also known as Mirk from minibrain-related kinase) and class II DYRKs, DYRK2, DYRK3 (also known as REDK from regulatory erythroid kinase) and DYRK4 (Figure 1A). DYRK kinases phosphorylate a broad set of substrates that are involved in a wide range of cellular processes, and they are thought to fulfill essential biological functions both during development and in maintaining homeostasis during the adult life [4,43,94,126,127]. Consequently, the aberrant regulation or expression of DYRK kinases has been associated with several human pathologies, including cancer.
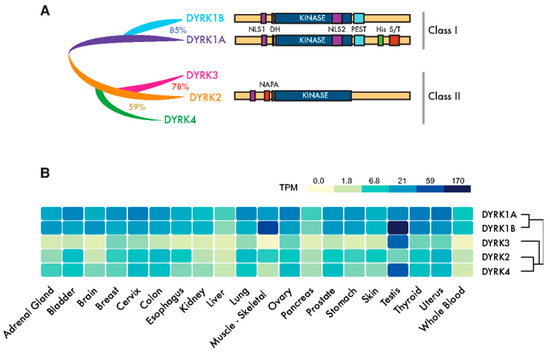
Figure 1. Dual-specificity tyrosine-regulated kinase (DYRK) protein kinases: primary structure and expression. (A) Scheme of the mammalian family of DYRKs, indicating their phylogenic relationships, degree of homology and protein domains. The catalytic domain (KINASE) and the DYRK homology box (DH) are common to all members of the family. Class I DYRKs have two nuclear localization signals (NLSs) (NLS1 and NLS2) and a proline-, glutamic acid-, serine- and threonine-rich (PEST) motif. DYRK1A also includes a tract of 13 consecutive histidine residues (His) and a region enriched in serine/threonine residues (S/T) at the C-terminus. Class II DYRKs have a common structure, with the characteristic N-terminal autophosphorylation accessory (NAPA) domain at the N-terminus. In the case of DYRK2 and DYRK4, functional NLSs have been described within the noncatalytic N-terminus. (B) The expression of human DYRKs based on the Genotype-Tissue Expression (GTEx) data represented as the median TPMs (transcripts per million: GTEx Analysis Release V8, www.gtexportal.org/home, dbGaP Accession phs000424.v8.p2; brain: cortex; cervix: ectocervix; colon: sigmoid colon; esophagus; mucosa, kidney: cortex; skin: suprapubic—not sun-exposed).
The members of the DYRK family all share a highly conserved catalytic domain with special features within the CMGC group [5] and the so-called DYRK homology (DH) box motif located upstream of it (Figure 1A). In addition, DYRK kinases present class-specific domains: DYRK1A and DYRK1B harbor a proline-, glutamic acid-, serine- and threonine-rich (PEST) motif in the noncatalytic C-terminal region and equally positioned nuclear localization signals (NLS) (Figure 1A). On the other hand, class II DYRKs present a N-terminal autophosphorylation accessory region (NAPA) domain, essential for catalytic activation [6] (Figure 1A). All human DYRKs accumulate in the cytosol of cells, and DYRK1A, DYRK2 and DYRK4 can be imported into the nucleus by means of dedicated NLSs [7][8][9]. Nuclear DYRK1A is a chromatin-associated kinase capable of regulating the gene expression [10][11] , and it is functionally linked to the DNA damage response (DDR) [12][13][14]. Chromatin association in the DDR context has also been described for DYRK1B [15]. Moreover, a DYRK1A-specific run of histidine residues targets this family member to the subnuclear splicing compartment [7], and the noncatalytic N-terminal domain of DYRK3 serves to localize it to stress granules [16]. Both the histidine run in DYRK1A and the N-terminus of DYRK3 participate in the generation of phase-separated subcellular compartments [17][18]. Changes in the subcellular localization of DYRK proteins have been observed in response to different signals, such as that of DYRK2 in response to DNA damage or proinflammatory signals [19][20] or DYRK1A in response to Wnt signaling [21]. However, how the subcellular localization of DYRKs is regulated or how it contributes to their activity is still not well-understood.
DYRK1A and DYRK1B are expressed ubiquitously in human tissues, whereas class II DYRKs are generally expressed more weakly and in a more tissue-restricted pattern (Figure 1B). The expression of DYRKs is regulated through alternative promoters that generate transcripts with distinct 5′-untranslated regions and/or encoding different N-terminal regions [4]. In addition, alternative splicing generates multiple protein isoforms of unclear functional significance [4][8][22][23][24]. DYRKs are also subject to other post-transcriptional events, such as microRNAs-mediated gene silencing [25][26][27] or local translation [28].
DYRK kinases are “dual specificity” kinases, as they can phosphorylate both tyrosine (Y) and serine/threonine (S/T) residues, although Y-phosphorylation is limited to their autophosphorylation activity [29]. These kinases are activated by the phosphorylation of residues within the activation loop, which drives a conformational switch from the inactive to active state [30][31]. Unlike other kinase families, this key event in DYRKs is an autocatalytic reaction that occurs during protein synthesis and that generates a constitutively active kinase [32]. As DYRK activation does not depend on upstream kinases, other regulatory mechanisms are thought to operate, including the dephosphorylation of residues in the activation loop, allosteric phosphorylation performed by other kinases [9][33][34][35][36], interactions with scaffolding proteins [37][38][39], accessibility to substrates due to changes in the subcellular localization or regulation of protein levels at transcriptional and/or post-transcriptional levels.
2. The Role of DYRKs in Cancer
DYRKs phosphorylate a wide range of substrates, including factors associated with one or several of the hallmarks of cancer [40] (Figure 2). Of all the DYRKs, only DYRK1A has been identified in high-throughput cancer studies, initially as a potential tumor suppressor using Tumor Suppressor and Oncogene Explorer (TUSON) [41], and later on, it was proposed as a driver in liver cancer through a study that identified such drivers according to mutations in unusual nucleotide contexts [42]. Although these results would suggest that DYRKs are not major drivers of cancer, further evidence that they play a role in oncogenic processes has emerged over the past two decades, either by detecting alterations to the DYRK expression in tumor tissues, and/or by analyzing the impact of DYRK-dependent phosphorylation on substrates involved in cancer-related events.
Figure 2. DYRKs are involved in cancer-associated processes. DYRK kinases participate in the regulation of crucial cell events, the perturbation of which is responsible for producing important features in cancer cells or the hallmarks of cancer.
DYRK1A is the most extensively studied member of the family, mainly due to its key role in neurogenesis and in the etiology of some of the pathological traits associated to Down syndrome (recently reviewed in [43]). In addition, DYRK1A haploinsufficiency caused by de novo truncations or by missense-inactivating mutations underlies a rare, severe disorder, the DYRK1A haploinsufficiency syndrome (also known as MRD7 or Mental Retardation, Autosomal Dominant 7: OMIM#614104 and ORPHA:464311 and 268261; [44][45] and references therein). Regarding a putative role in cancer, several studies have ascribed opposite functions to this family member [46][47][48][49][50], reflecting a very complex scenario. Therefore, it remains unclear as to whether DYRK1A acts as a tumor suppressor or a tumor promoter or, more probably, as either, depending on the tumor context. Unlike DYRK1A, findings on its closest paralog DYRK1B point mostly to a prosurvival and protumorigenic role (recently reviewed in [51]). DYRK2 activity appears to affect crucial processes linked to tumor progression like the cell cycle, the DDR, epithelial-to-mesenchymal transition, the xenobiotic response system and cellular proteostasis (recently reviewed in [52][53]). Alterations in the expression of DYRK2 have been observed in different types of cancer, and in general, the weaker the expression of DYRK2, the worse the prognosis [54][55][56][57][58][59][60][61]; in addition, DYRK2 gene silencing confers an enhanced proliferative capacity and metastatic potential in vivo [56][61][62]. However, some discordant phenotypes have been described when studying breast cancer [63][64][65][66]. Finally, the contribution of DYRK3 and DYRK4 to tumorigenesis is less clear, with very little evidence for the participation of DYRK3 and no evidence for that of DYRK4.
3. DYRK Inhibitors as Antitumor Therapies
Chemical compounds that bind and functionally block protein kinases have been studied extensively and employed as antitumor agents, both in research and in clinical trials. Although the role of DYRK family members in tumorigenesis and tumor progression has not been fully elucidated, pharmacological inhibitors of DYRK kinases have been tested in laboratories for their antimalignant activity, and a few of them are already undergoing clinical trials.
In the case of DYRK1A, the search for both naturally occurring and synthetic inhibitors has been extensive given that DYRK1A may be a potential pharmacological target not only in cancer but, also, in neurodegenerative diseases (reviewed in [43]), Down syndrome [67][68][69][70] and diabetes (reviewed in [71]). Comprehensive reviews on DYRK1A inhibitors can be found at [72][73][74]. Compounds targeting DYRK1B, with either restricted or broad specificity, have been used as research tools, and they display toxicity towards several types of cancer cells or they promote the cell cycle re-entry of quiescent tumor cells (reviewed in [51]), with positive effects in combinatorial drug approaches. For instance, the DYRK1B inhibitor AZ191 [75] increases the anticancer effects of doxorubicin in liposarcoma cell lines [76] or sensitizes pancreatic adenocarcinoma cell lines to mTOR inhibition [77]. However, AZ191 has been also shown to counteract the antitumor effects of the lysosome inhibitor Bafilomycin A1 in hepatocellular carcinoma cell lines [78]. For DYRK2, experimental data on the antitumor effects of the natural DYRK2 inhibitor curcumin and of the synthetic compound LDN192960 was obtained in both in vitro and in vivo models of triple negative breast cancer (TNBC) and multiple myeloma, supporting the hypothesis that DYRK2 is a promising pharmaceutical target in these malignancies [64][79]. Finally, better understanding the role of DYRKs in tumor cells has proven valuable by helping to identify combinatorial therapeutic approaches, as in the cases of the DYRK1B inhibitors that enhance the inhibitory efficiency of MEK and mTOR [80][81][82] or DYRK2 inhibition sensitizing MDA-MB-468 cells to the proteasome inhibitor bortezomib [66].
The only inhibitors of the DYRK family members currently being screened in clinical trials were identified as inhibitors of other protein kinases. In particular, compound CX-4945 was initially identified as a casein kinase 2 inhibitor, but it was subsequently shown to be a potent DYRK1A and DYRK1B inhibitor [69], and it is currently in phase I and II clinical studies for medulloblastoma, cholangiocarcinoma and basal cell carcinoma (NCT02128282, NCT03904862 and NCT03897036). Compound OTS167, a chemical initially described as a maternal embryonic leucine zipper kinase inhibitor, has potent anti-DYRK1A activity [83], and it is currently being assessed in clinical trials for the treatment of advanced breast cancer and TNBC (phase I) and for multiple types of leukemia (phase II: NCT02795520). Finally, two other DYRK inhibitors have been assessed in clinical trials for non-neoplastic disorders: GSK-626616 [16] completed a phase I clinical trial to evaluate its action on anemia (NCT00443170), and lorecivivint, a potent CLK2 inhibitor that also inhibits DYRK1A [84], is being studied in a phase II trial for the treatment of moderate-to-severe symptomatic osteoarthritis (NCT03706521). Thus, they could be repurposed in trials for the treatment of specific cancer types.
4. Conclusions
In the last decade, more experimental evidence indicates that DYRK protein kinases are a novel class of “kinase-of-interest” in cancer. However, this evidence mostly comes from studies exploring DYRK expressions in tumor tissues and/or the phenotypic changes triggered by manipulating the DYRK protein levels in cancer cell lines. These data not only provide a partial and confusing picture of the influence of DYRKs in tumor initiation and progression, but also, they highlight the many questions that still need to be addressed. In particular, it remains unclear which molecular pathways are regulated by DYRKs in different tumor types and which of them selectively trigger cells to engage in neoplastic transformation or enhance the malignant phenotype of tumor cells. Resolving these issues will not only help understand the biology behind the activity of these kinases, but also, it will provide a basis for the rational design of therapeutic approaches based on inhibitors. In this regard, while incomplete, the currently available data provides precious information on which forthcoming therapeutic approaches may be based. Therefore, the tumor types in which downregulation of the DYRK kinase has been associated with increased tumor growth and/or invasiveness should not be considered for trials with DYRK inhibitors. Conversely, inhibitors targeting DYRK family members that are known to favor the tumorigenesis of specific tumor types should be considered for such trials. Nevertheless, putative side effects due to the inhibition of members that are essential to maintaining cellular homeostasis in normal cells, such as the dosage-sensitive DYRK1A or DYRK1B, should be carefully monitored. In this context, engineering drugs to increase their specificity, exclusively targeting proliferating cells, would be worthwhile. Finally, and considering the differential and sometimes opposite roles of distinct DYRK kinases in tumor progression, selectivity towards a specific member of the family is crucial and, at the same time, very challenging, particularly given the strong structural similarity of the catalytic domain. Smart solutions might include an allosteric drug design or other additional efforts to increase compound selectivity.
To conclude, many important advances in understanding how the dysregulation of DYRK protein kinases is associated to pathological phenotypes in humans have been made in recent years—in particular, in terms of their involvement in cancer. Still, many secrets behind the oncogenic or protective potential of DYRK kinases remain to be revealed, and we anticipate that the field will continue to grow for the foreseeable future.
This entry is adapted from the peer-reviewed paper 10.3390/cancers12082106
References
- Collett, M.S.; Erikson, R.L. Protein kinase activity associated with the avian sarcoma virus src gene product. Proc. Natl. Acad. Sci. USA 1978, 75, 2021–2024, doi:10.1073/pnas.75.4.2021.
- Fleuren, E.D.; Zhang, L.; Wu, J.; Daly, R.J. The kinome ‘at large’ in cancer. Nat. Rev. Cancer 2016, 16, 83–98, doi:10.1038/nrc.2015.18.
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934, doi:10.1126/science.1075762.
- Aranda, S.; Laguna, A.; de la Luna, S. DYRK family of protein kinases: Evolutionary relationships, biochemical properties, and functional roles. FASEB J. 2011, 25, 449–462, doi:10.1096/fj.10-165837.
- Kannan, N.; Neuwald, A.F. Evolutionary constraints associated with functional specificity of the CMGC protein kinases MAPK, CDK, GSK, SRPK, DYRK, and CK2alpha. Protein Sci. 2004, 13, 2059–2077.
- Kinstrie, R.; Luebbering, N.; Miranda-Saavedra, D.; Sibbet, G.; Han, J.; Lochhead, P.A.; Cleghon, V. Characterization of a domain that transiently converts class 2 DYRKs into intramolecular tyrosine kinases. Sci. Signal. 2010, 3, ra16, doi:10.1126/scisignal.2000579.
- Alvarez, M.; Estivill, X.; de la Luna, S. DYRK1A accumulates in splicing speckles through a novel targeting signal and induces speckle disassembly. J. Cell Sci. 2003, 116, 3099–3107, doi:10.1242/jcs.00618.
- Papadopoulos, C.; Arato, K.; Lilienthal, E.; Zerweck, J.; Schutkowski, M.; Chatain, N.; Muller-Newen, G.; Becker, W.; de la Luna, S. Splice variants of the dual specificity tyrosine phosphorylation-regulated kinase 4 (DYRK4) differ in their subcellular localization and catalytic activity. J. Biol. Chem. 2011, 286, 5494–5505, doi:10.1074/jbc.M110.157909.
- Taira, N.; Yamamoto, H.; Yamaguchi, T.; Miki, Y.; Yoshida, K. ATM augments nuclear stabilization of DYRK2 by inhibiting MDM2 in the apoptotic response to DNA damage. J. Biol. Chem. 2010, 285, 4909–4919, doi:10.1074/jbc.M109.042341.
- Di Vona, C.; Bezdan, D.; Islam, A.B.; Salichs, E.; Lopez-Bigas, N.; Ossowski, S.; de la Luna, S. Chromatin-wide profiling of DYRK1A reveals a role as a gene-specific RNA polymerase II CTD kinase. Mol. Cell 2015, 57, 506–520, doi:10.1016/j.molcel.2014.12.026.
- Yu, D.; Cattoglio, C.; Xue, Y.; Zhou, Q. A complex between DYRK1A and DCAF7 phosphorylates the C-terminal domain of RNA polymerase II to promote myogenesis. Nucleic Acids Res. 2019, 47, 4462–4475, doi:10.1093/nar/gkz162.
- Roewenstrunk, J.; Di Vona, C.; Chen, J.; Borras, E.; Dong, C.; Arato, K.; Sabido, E.; Huen, M.S.Y.; de la Luna, S. A comprehensive proteomics-based interaction screen that links DYRK1A to RNF169 and to the DNA damage response. Sci. Rep. 2019, 9, 6014, doi:10.1038/s41598-019-42445-x.
- Menon, V.R.; Ananthapadmanabhan, V.; Swanson, S.; Saini, S.; Sesay, F.; Yakovlev, V.; Florens, L.; DeCaprio, J.A.; Washburn, M.P.; Dozmorov, M.; et al. DYRK1A regulates the recruitment of 53BP1 to the sites of DNA damage in part through interaction with RNF169. Cell Cycle 2019, 18, 531–551, doi:10.1080/15384101.2019.1577525.
- Guard, S.E.; Poss, Z.C.; Ebmeier, C.C.; Pagratis, M.; Simpson, H.; Taatjes, D.J.; Old, W.M. The nuclear interactome of DYRK1A reveals a functional role in DNA damage repair. Sci. Rep. 2019, 9, 6539, doi:10.1038/s41598-019-42990-5.
- Dong, C.; West, K.L.; Tan, X.Y.; Li, J.; Ishibashi, T.; Yu, C.H.; Sy, S.M.H.; Leung, J.W.C.; Huen, M.S.Y. Screen identifies DYRK1B network as mediator of transcription repression on damaged chromatin. Proc. Natl. Acad. Sci. USA 2020, doi:10.1073/pnas.2002193117.
- Wippich, F.; Bodenmiller, B.; Trajkovska, M.G.; Wanka, S.; Aebersold, R.; Pelkmans, L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 2013, 152, 791–805, doi:10.1016/j.cell.2013.01.033.
- Rai, A.K.; Chen, J.X.; Selbach, M.; Pelkmans, L. Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 2018, 559, 211–216, doi:10.1038/s41586-018-0279-8.
- Lu, H.; Yu, D.; Hansen, A.S.; Ganguly, S.; Liu, R.; Heckert, A.; Darzacq, X.; Zhou, Q. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 2018, 558, 318–323, doi:10.1038/s41586-018-0174-3.
- Taira, N.; Nihira, K.; Yamaguchi, T.; Miki, Y.; Yoshida, K. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol. Cell 2007, 25, 725–738, doi:10.1016/j.molcel.2007.02.007.
- Xu, L.; Sun, Y.; Li, M.; Ge, X. Dyrk2 mediated the release of proinflammatory cytokines in LPS-induced BV2 cells. Int. J. Biol. Macromol. 2018, 109, 1115–1124, doi:10.1016/j.ijbiomac.2017.11.095.
- Granno, S.; Nixon-Abell, J.; Berwick, D.C.; Tosh, J.; Heaton, G.; Almudimeegh, S.; Nagda, Z.; Rain, J.C.; Zanda, M.; Plagnol, V.; et al. Downregulated Wnt/beta-catenin signalling in the Down syndrome hippocampus. Sci. Rep. 2019, 9, 7322, doi:10.1038/s41598-019-43820-4.
- Guimera, J.; Casas, C.; Estivill, X.; Pritchard, M. Human minibrain homologue (MNBH/DYRK1): Characterization, alternative splicing, differential tissue expression, and overexpression in Down syndrome. Genomics 1999, 57, 407–418, doi:10.1006/geno.1999.5775.
- Leder, S.; Weber, Y.; Altafaj, X.; Estivill, X.; Joost, H.G.; Becker, W. Cloning and characterization of DYRK1B, a novel member of the DYRK family of protein kinases. Biochem. Biophys. Res. Commun. 1999, 254, 474–479, doi:10.1006/bbrc.1998.9967.
- Lord, K.A.; Creasy, C.L.; King, A.G.; King, C.; Burns, B.M.; Lee, J.C.; Dillon, S.B. REDK, a novel human regulatory erythroid kinase. Blood 2000, 95, 2838–2846.
- da Costa Martins, P.A.; Salic, K.; Gladka, M.M.; Armand, A.S.; Leptidis, S.; el Azzouzi, H.; Hansen, A.; Coenen-de Roo, C.J.; Bierhuizen, M.F.; van der Nagel, R.; et al. MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nat. Cell Biol. 2010, 12, 1220–1227, doi:10.1038/ncb2126.
- Zhang, Y.; Liao, J.M.; Zeng, S.X.; Lu, H. p53 downregulates Down syndrome-associated DYRK1A through miR-1246. EMBO Rep. 2011, 12, 811–817, doi:10.1038/embor.2011.98.
- Kim, J.; Siverly, A.N.; Chen, D.; Wang, M.; Yuan, Y.; Wang, Y.; Lee, H.; Zhang, J.; Muller, W.J.; Liang, H.; et al. Ablation of miR-10b suppresses oncogene-induced mammary tumorigenesis and metastasis and reactivates tumor-suppressive pathways. Cancer Res. 2016, 76, 6424–6435, doi:10.1158/0008-5472.CAN-16-1571.
- Vidaki, M.; Drees, F.; Saxena, T.; Lanslots, E.; Taliaferro, M.J.; Tatarakis, A.; Burge, C.B.; Wang, E.T.; Gertler, F.B. A requirement for Mena, an actin regulator, in local mRNA translation in developing neurons. Neuron 2017, 95, 608–622, doi:10.1016/j.neuron.2017.06.048.
- Himpel, S.; Panzer, P.; Eirmbter, K.; Czajkowska, H.; Sayed, M.; Packman, L.C.; Blundell, T.; Kentrup, H.; Grotzinger, J.; Joost, H.G.; et al. Identification of the autophosphorylation sites and characterization of their effects in the protein kinase DYRK1A. Biochem. J. 2001, 359, 497–505.
- Kentrup, H.; Becker, W.; Heukelbach, J.; Wilmes, A.; Schurmann, A.; Huppertz, C.; Kainulainen, H.; Joost, H.G. Dyrk, a dual specificity protein kinase with unique structural features whose activity is dependent on tyrosine residues between subdomains VII and VIII. J. Biol. Chem. 1996, 271, 3488–3495, doi:10.1074/jbc.271.7.3488.
- Soundararajan, M.; Roos, A.K.; Savitsky, P.; Filippakopoulos, P.; Kettenbach, A.N.; Olsen, J.V.; Gerber, S.A.; Eswaran, J.; Knapp, S.; Elkins, J.M. Structures of Down syndrome kinases, DYRKs, reveal mechanisms of kinase activation and substrate recognition. Structure 2013, 21, 986–996, doi:10.1016/j.str.2013.03.012.
- Lochhead, P.A.; Sibbet, G.; Morrice, N.; Cleghon, V. Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell 2005, 121, 925–936, doi:10.1016/j.cell.2005.03.034.
- Lim, S.; Zou, Y.; Friedman, E. The transcriptional activator Mirk/Dyrk1B is sequestered by p38alpha/beta MAP kinase. J. Biol. Chem. 2002, 277, 49438–49445, doi:10.1074/jbc.M206840200.
- Varjosalo, M.; Bjorklund, M.; Cheng, F.; Syvanen, H.; Kivioja, T.; Kilpinen, S.; Sun, Z.; Kallioniemi, O.; Stunnenberg, H.G.; He, W.W.; et al. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell 2008, 133, 537–548, doi:10.1016/j.cell.2008.02.047.
- Tschop, K.; Conery, A.R.; Litovchick, L.; Decaprio, J.A.; Settleman, J.; Harlow, E.; Dyson, N. A kinase shRNA screen links LATS2 and the pRB tumor suppressor. Genes Dev. 2011, 25, 814–830, doi:10.1101/gad.2000211.
- Ashford, A.L.; Dunkley, T.P.; Cockerill, M.; Rowlinson, R.A.; Baak, L.M.; Gallo, R.; Balmanno, K.; Goodwin, L.M.; Ward, R.A.; Lochhead, P.A.; et al. Identification of DYRK1B as a substrate of ERK1/2 and characterisation of the kinase activity of DYRK1B mutants from cancer and metabolic syndrome. Cell Mol. Life Sci. 2016, 73, 883–900, doi:10.1007/s00018-015-2032-x.
- Alvarez, M.; Altafaj, X.; Aranda, S.; de la Luna, S. DYRK1A autophosphorylation on serine residue 520 modulates its kinase activity via 14-3-3 binding. Mol. Biol. Cell 2007, 18, 1167–1178, doi:10.1091/mbc.E06-08-0668.
- Maddika, S.; Chen, J. Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat. Cell Biol. 2009, 11, 409–419, doi:10.1038/ncb1848.
- Glenewinkel, F.; Cohen, M.J.; King, C.R.; Kaspar, S.; Bamberg-Lemper, S.; Mymryk, J.S.; Becker, W. The adaptor protein DCAF7 mediates the interaction of the adenovirus E1A oncoprotein with the protein kinases DYRK1A and HIPK2. Sci. Rep. 2016, 6, 28241, doi:10.1038/srep28241.
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674, doi:10.1016/j.cell.2011.02.013.
- Davoli, T.; Xu, A.W.; Mengwasser, K.E.; Sack, L.M.; Yoon, J.C.; Park, P.J.; Elledge, S.J. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 2013, 155, 948–962, doi:10.1016/j.cell.2013.10.011.
- Dietlein, F.; Weghorn, D.; Taylor-Weiner, A.; Richters, A.; Reardon, B.; Liu, D.; Lander, E.S.; Van Allen, E.M.; Sunyaev, S.R. Identification of cancer driver genes based on nucleotide context. Nat. Genet. 2020, 52, 208–218, doi:10.1038/s41588-019-0572-y.
- Arbones, M.L.; Thomazeau, A.; Nakano-Kobayashi, A.; Hagiwara, M.; Delabar, J.M. DYRK1A and cognition: A lifelong relationship. Pharmacol. Ther. 2019, 194, 199–221, doi:10.1016/j.pharmthera.2018.09.010.
- van Bon, B.W.; Coe, B.P.; Bernier, R.; Green, C.; Gerdts, J.; Witherspoon, K.; Kleefstra, T.; Willemsen, M.H.; Kumar, R.; Bosco, P.; et al. Disruptive de novo mutations of DYRK1A lead to a syndromic form of autism and ID. Mol. Psychiatry 2016, 21, 126–132, doi:10.1038/mp.2015.5.
- Arranz, J.; Balducci, E.; Arato, K.; Sanchez-Elexpuru, G.; Najas, S.; Parras, A.; Rebollo, E.; Pijuan, I.; Erb, I.; Verde, G.; et al. Impaired development of neocortical circuits contributes to the neurological alterations in DYRK1A haploinsufficiency syndrome. Neurobiol. Dis. 2019, 127, 210–222, doi:10.1016/j.nbd.2019.02.022.
- Liu, Q.; Liu, N.; Zang, S.; Liu, H.; Wang, P.; Ji, C.; Sun, X. Tumor suppressor DYRK1A effects on proliferation and chemoresistance of AML cells by downregulating c-Myc. PLoS ONE 2014, 9, e98853, doi:10.1371/journal.pone.0098853.
- Pozo, N.; Zahonero, C.; Fernandez, P.; Linares, J.M.; Ayuso, A.; Hagiwara, M.; Perez, A.; Ricoy, J.R.; Hernandez-Lain, A.; Sepulveda, J.M.; et al. Inhibition of DYRK1A destabilizes EGFR and reduces EGFR-dependent glioblastoma growth. J. Clin. Investig. 2013, 123, 2475–2487, doi:10.1172/JCI63623.
- Luna, J.; Boni, J.; Cuatrecasas, M.; Bofill-De Ros, X.; Nunez-Manchon, E.; Gironella, M.; Vaquero, E.C.; Arbones, M.L.; de la Luna, S.; Fillat, C. DYRK1A modulates c-MET in pancreatic ductal adenocarcinoma to drive tumour growth. Gut 2019, 68, 1465–1476, doi:10.1136/gutjnl-2018-316128.
- MacDonald, J.; Ramos-Valdes, Y.; Perampalam, P.; Litovchick, L.; DiMattia, G.E.; Dick, F.A. A systematic analysis of negative growth control implicates the DREAM complex in cancer cell dormancy. Mol. Cancer Res. 2017, 15, 371–381, doi:10.1158/1541-7786.MCR-16-0323-T.
- Radhakrishnan, A.; Nanjappa, V.; Raja, R.; Sathe, G.; Puttamallesh, V.N.; Jain, A.P.; Pinto, S.M.; Balaji, S.A.; Chavan, S.; Sahasrabuddhe, N.A.; et al. A dual specificity kinase, DYRK1A, as a potential therapeutic target for head and neck squamous cell carcinoma. Sci. Rep. 2016, 6, 36132, doi:10.1038/srep36132.
- Becker, W. A wake-up call to quiescent cancer cells—potential use of DYRK1B inhibitors in cancer therapy. FEBS J. 2018, 285, 1203–1211, doi:10.1111/febs.14347.
- Yoshida, S.; Yoshida, K. Multiple functions of DYRK2 in cancer and tissue development. FEBS Lett. 2019, 593, 2953–2965, doi:10.1002/1873-3468.13601.
- Correa-Saez, A.; Jimenez-Izquierdo, R.; Garrido-Rodriguez, M.; Morrugares, R.; Munoz, E.; Calzado, M.A. Updating dual-specificity tyrosine-phosphorylation-regulated kinase 2 (DYRK2): Molecular basis, functions and role in diseases. Cell Mol. Life Sci. 2020, doi:10.1007/s00018-020-03556-1.
- Yan, H.; Hu, K.; Wu, W.; Li, Y.; Tian, H.; Chu, Z.; Koeffler, H.P.; Yin, D. Low expression of DYRK2 (Dual Specificity Tyrosine Phosphorylation Regulated Kinase 2) correlates with poor prognosis in colorectal cancer. PLoS ONE 2016, 11, e0159954, doi:10.1371/journal.pone.0159954.
- Zhang, X.; Xu, P.; Ni, W.; Fan, H.; Xu, J.; Chen, Y.; Huang, W.; Lu, S.; Liang, L.; Liu, J.; et al. Downregulated DYRK2 expression is associated with poor prognosis and Oxaliplatin resistance in hepatocellular carcinoma. Pathol. Res. Pract. 2016, 212, 162–170, doi:10.1016/j.prp.2016.01.002.
- Yokoyama-Mashima, S.; Yogosawa, S.; Kanegae, Y.; Hirooka, S.; Yoshida, S.; Horiuchi, T.; Ohashi, T.; Yanaga, K.; Saruta, M.; Oikawa, T.; et al. Forced expression of DYRK2 exerts anti-tumor effects via apoptotic induction in liver cancer. Cancer Lett. 2019, 451, 100–109, doi:10.1016/j.canlet.2019.02.046.
- Shen, Y.; Zhang, L.; Wang, D.; Bao, Y.; Liu, C.; Xu, Z.; Huang, W.; Cheng, C. Regulation of glioma cells migration by DYRK2. Neurochem. Res. 2017, 42, 3093–3102, doi:10.1007/s11064-017-2345-2.
- Yamashita, S.; Chujo, M.; Tokuishi, K.; Anami, K.; Miyawaki, M.; Yamamoto, S.; Kawahara, K. Expression of dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 2 (DYRK2) can be a favorable prognostic marker in pulmonary adenocarcinoma. J. Thorac. Cardiovasc. Surg. 2009, 138, 1303–1308, doi:10.1016/j.jtcvs.2009.08.003.
- Yamashita, S.; Chujo, M.; Moroga, T.; Anami, K.; Tokuishi, K.; Miyawaki, M.; Kawano, Y.; Takeno, S.; Yamamoto, S.; Kawahara, K. DYRK2 expression may be a predictive marker for chemotherapy in non-small cell lung cancer. Anticancer Res. 2009, 29, 2753–2757.
- Nomura, S.; Suzuki, Y.; Takahashi, R.; Terasaki, M.; Kimata, R.; Terasaki, Y.; Hamasaki, T.; Kimura, G.; Shimizu, A.; Kondo, Y. Dual-specificity tyrosine phosphorylation-regulated kinase 2 (DYRK2) as a novel marker in T1 high-grade and T2 bladder cancer patients receiving neoadjuvant chemotherapy. BMC Urol. 2015, 15, 53, doi:10.1186/s12894-015-0040-7.
- Ito, D.; Yogosawa, S.; Mimoto, R.; Hirooka, S.; Horiuchi, T.; Eto, K.; Yanaga, K.; Yoshida, K. Dual-specificity tyrosine-regulated kinase 2 is a suppressor and potential prognostic marker for liver metastasis of colorectal cancer. Cancer Sci. 2017, 108, 1565–1573, doi:10.1111/cas.13280.
- Yamaguchi, N.; Mimoto, R.; Yanaihara, N.; Imawari, Y.; Hirooka, S.; Okamoto, A.; Yoshida, K. DYRK2 regulates epithelial-mesenchymal-transition and chemosensitivity through Snail degradation in ovarian serous adenocarcinoma. Tumour Biol. 2015, 36, 5913–5923, doi:10.1007/s13277-015-3264-y.
- Taira, N.; Mimoto, R.; Kurata, M.; Yamaguchi, T.; Kitagawa, M.; Miki, Y.; Yoshida, K. DYRK2 priming phosphorylation of c-Jun and c-Myc modulates cell cycle progression in human cancer cells. J. Clin. Investig. 2012, 122, 859–872, doi:60818 [pii]10.1172/JCI60818.
- Banerjee, S.; Wei, T.; Wang, J.; Lee, J.J.; Gutierrez, H.L.; Chapman, O.; Wiley, S.E.; Mayfield, J.E.; Tandon, V.; Juarez, E.F.; et al. Inhibition of dual-specificity tyrosine phosphorylation-regulated kinase 2 perturbs 26S proteasome-addicted neoplastic progression. Proc. Natl. Acad. Sci. USA 2019, 116, 24881–24891, doi:10.1073/pnas.1912033116.
- Enomoto, Y.; Yamashita, S.; Yoshinaga, Y.; Fukami, Y.; Miyahara, S.; Nabeshima, K.; Iwasaki, A. Downregulation of DYRK2 can be a predictor of recurrence in early stage breast cancer. Tumour Biol. 2014, 35, 11021–11025, doi:10.1007/s13277-014-2413-z.
- Guo, X.; Wang, X.; Wang, Z.; Banerjee, S.; Yang, J.; Huang, L.; Dixon, J.E. Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nat. Cell Biol. 2016, 18, 202–212, doi:10.1038/ncb3289.
- Nguyen, T.L.; Duchon, A.; Manousopoulou, A.; Loaec, N.; Villiers, B.; Pani, G.; Karatas, M.; Mechling, A.E.; Harsan, L.A.; Limanton, E.; et al. Correction of cognitive deficits in mouse models of Down syndrome by a pharmacological inhibitor of DYRK1A. Dis. Model. Mech. 2018, 11, doi:10.1242/dmm.035634.
- Neumann, F.; Gourdain, S.; Albac, C.; Dekker, A.D.; Bui, L.C.; Dairou, J.; Schmitz-Afonso, I.; Hue, N.; Rodrigues-Lima, F.; Delabar, J.M.; et al. DYRK1A inhibition and cognitive rescue in a Down syndrome mouse model are induced by new fluoro-DANDY derivatives. Sci. Rep. 2018, 8, 2859, doi:10.1038/s41598-018-20984-z.
- Kim, H.; Lee, K.S.; Kim, A.K.; Choi, M.; Choi, K.; Kang, M.; Chi, S.W.; Lee, M.S.; Lee, J.S.; Lee, S.Y.; et al. A chemical with proven clinical safety rescues Down-syndrome-related phenotypes in through DYRK1A inhibition. Dis. Model. Mech. 2016, 9, 839–848, doi:10.1242/dmm.025668.
- de la Torre, R.; de Sola, S.; Hernandez, G.; Farre, M.; Pujol, J.; Rodriguez, J.; Espadaler, J.M.; Langohr, K.; Cuenca-Royo, A.; Principe, A.; et al. Safety and efficacy of cognitive training plus epigallocatechin-3-gallate in young adults with Down’s syndrome (TESDAD): A double-blind, randomised, placebo-controlled, phase 2 trial. Lancet Neurol. 2016, 15, 801–810, doi:10.1016/S1474-4422(16)30034-5.
- Belgardt, B.F.; Lammert, E. DYRK1A: A promising drug target for islet transplant-based Diabetes therapies. Diabetes 2016, 65, 1496–1498, doi:10.2337/dbi16-0013.
- Ionescu, A.; Dufrasne, F.; Gelbcke, M.; Jabin, I.; Kiss, R.; Lamoral-Theys, D. DYRK1A kinase inhibitors with emphasis on cancer. Mini Rev. Med. Chem. 2012, 12, 1315–1329.
- Jarhad, D.B.; Mashelkar, K.K.; Kim, H.R.; Noh, M.; Jeong, L.S. Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase 1A (DYRK1A) inhibitors as potential therapeutics. J. Med. Chem. 2018, 61, 9791–9810, doi:10.1021/acs.jmedchem.8b00185.
- Nguyen, T.L.; Fruit, C.; Herault, Y.; Meijer, L.; Besson, T. Dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) inhibitors: A survey of recent patent literature. Expert Opin. Ther. Pat. 2017, 27, 1183–1199, doi:10.1080/13543776.2017.1360285.
- Ashford, A.L.; Oxley, D.; Kettle, J.; Hudson, K.; Guichard, S.; Cook, S.J.; Lochhead, P.A. A novel DYRK1B inhibitor AZ191 demonstrates that DYRK1B acts independently of GSK3beta to phosphorylate cyclin D1 at Thr (286), not Thr (288). Biochem. J. 2014, 457, 43–56, doi:10.1042/BJ20130461.
- Chen, H.; Shen, J.; Choy, E.; Hornicek, F.J.; Shan, A.; Duan, Z. Targeting DYRK1B suppresses the proliferation and migration of liposarcoma cells. Oncotarget 2018, 9, 13154–13166, doi:10.18632/oncotarget.22743.
- Singh, R.; Dhanyamraju, P.K.; Lauth, M. DYRK1B blocks canonical and promotes non-canonical Hedgehog signaling through activation of the mTOR/AKT pathway. Oncotarget 2017, 8, 833–845, doi:10.18632/oncotarget.13662.
- Yan, Y.; Jiang, K.; Liu, P.; Zhang, X.; Dong, X.; Gao, J.; Liu, Q.; Barr, M.P.; Zhang, Q.; Hou, X.; et al. Bafilomycin A1 induces caspase-independent cell death in hepatocellular carcinoma cells via targeting of autophagy and MAPK pathways. Sci. Rep. 2016, 6, 37052, doi:10.1038/srep37052
- Banerjee, S.; Ji, C.; Mayfield, J.E.; Goel, A.; Xiao, J.; Dixon, J.E.; Guo, X. Ancient drug curcumin impedes 26S proteasome activity by direct inhibition of dual-specificity tyrosine-regulated kinase 2. Proc. Natl. Acad. Sci. USA 2018, 115, 8155–8160, doi:10.1073/pnas.1806797115.
- Gao, J.; Zhao, Y.; Lv, Y.; Chen, Y.; Wei, B.; Tian, J.; Yang, Z.; Kong, F.; Pang, J.; Liu, J.L.; et al. Mirk/Dyrk1B mediates G0/G1 to S phase cell cycle progression and cell survival involving MAPK/ERK signaling in human cancer cells. Cancer Cell Int. 2013, 13, 2, doi:10.1186/1475-2867-13-2.
- Deng, X.; Hu, J.; Ewton, D.Z.; Friedman, E. Mirk/dyrk1B kinase is upregulated following inhibition of mTOR. Carcinogenesis 2014, 35, 1968–1976, doi:10.1093/carcin/bgu058.
- Kettle, J.G.; Ballard, P.; Bardelle, C.; Cockerill, M.; Colclough, N.; Critchlow, S.E.; Debreczeni, J.; Fairley, G.; Fillery, S.; Graham, M.A.; et al. Discovery and optimization of a novel series of Dyrk1B kinase inhibitors to explore a MEK resistance hypothesis. J. Med. Chem. 2015, 58, 2834–2844, doi:10.1021/acs.jmedchem.5b00098.
- Allegretti, P.A.; Horton, T.M.; Abdolazimi, Y.; Moeller, H.P.; Yeh, B.; Caffet, M.; Michel, G.; Smith, M.; Annes, J.P. Generation of highly potent DYRK1A-dependent inducers of human beta-cell replication via multi-dimensional compound optimization. Bioorg. Med. Chem. 2019, 115193, doi:10.1016/j.bmc.2019.115193.
- Deshmukh, V.; O’Green, A.L.; Bossard, C.; Seo, T.; Lamangan, L.; Ibanez, M.; Ghias, A.; Lai, C.; Do, L.; Cho, S.; et al. Modulation of the Wnt pathway through inhibition of CLK2 and DYRK1A by lorecivivint as a novel, potentially disease-modifying approach for knee osteoarthritis treatment. Osteoarthr. Cartil. 2019, 27, 1347–1360, doi:10.1016/j.joca.2019.05.006.
