Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biophysics
Plant natriuretic peptides (PNPs) are hormones that have been extracted from many different species, with the Arabidopsis thaliana PNP (AtPNP-A) being the most studied among them. AtPNP-A is a signaling molecule that consists of 130 residues and is secreted into the apoplast, under conditions of biotic or abiotic stress. AtPNP-A has distant sequence homology with human ANP, a protein that forms amyloid fibrils in vivo.
- Arabidopsis thaliana
- amyloid fibrils
- natriuretic peptides
- plant natriuretic peptides
- functional amyloid
1. Introduction
Amyloid fibrils are formed by proteins or peptides, that under certain conditions self-assemble into characteristic fibrillar structures [1]. These highly ordered structures are characterized by extreme stability, while conflicting evidence has emerged about the ability of proteases to fragment them [2][3]. Amyloid forming proteins do not share any similarity in sequence or native structure, albeit amyloid fibrils are characterized by a common structure, known as “cross-β” structure. In “cross-β” conformation, hydrogen-bonded β-strands are organized perpendicularly to the fibril axis shaping β-sheets, which are, in turn, organized parallel to the main axis of the fibril [4]. Amyloid fibrils can be characterized either as pathological or functional; the deposition of amyloid fibrils is the main hallmark of a group of conformational disorders, known as amyloidoses [5], while on the other hand, many organisms, ranging from bacteria to humans, exploit the properties of amyloid fibrils to perform physiological functions [6][7].
In humans, the major hallmark of isolated atrial amyloidosis (IAA) is the formation of fibrillar deposits in the atria of the aging heart. Their primary component is the atrial natriuretic peptide (ANP) [8], a small hormone that belongs to the family of natriuretic peptides (NPs) and consists of 28 amino acid residues [9]. Nevertheless, immunohistochemical studies have shown that other NPs are also present on these deposits, such as brain NP (BNP) [10] and proANP1–98 [11]. ANP regulates blood volume and pressure in the circulatory system, via binding to a cell surface receptor, namely the natriuretic peptide receptor-A (NPR-A) [12][13]. In 1991, Vesely and Giordano also discovered the existence of the NPs system in plants, after using antibodies against human ANP (hANP) in plant tissue extracts [14]. Studies supported this groundbreaking finding, and further expanded on the role of plant natriuretic peptides (PNPs) [15][16][17][18] suggesting that they actively contribute to protoplast cell volume regulation [19][20][21] and to the ion balance of plants [22]. Impressively, PNPs act upon binding to cell membrane receptors in the same fashion as hANP indicating that they activate similar signaling pathways [23][24][25].
One of the most studied members of the PNP family is the Arabidopsis thaliana PNP (AtPNP-A). AtPNP-A is a signaling molecule that is secreted into the apoplast under conditions of biotic and abiotic stress in order to regulate cell response, thus preserving plant homeostasis [26][27][28]. The pre-processed sequence of AtPNP-A consists of 130 residues and contains a 25-residue-long N-terminal signal peptide which is critical for its secretion into the apoplast [28][29]. The AtPNP-A sequence contains a 34 amino acid region (AtPNP-A36–69), which is pivotal for its biological activity [21] (Figure 1). This peptide was also found to be conserved among all PNPs [20].
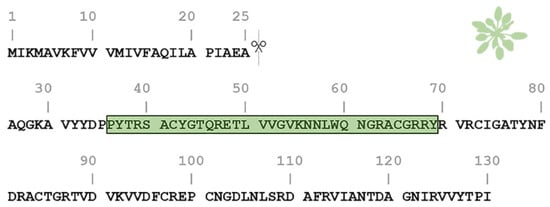
Figure 1. Amino acid sequence of the Arabidopsis thaliana plant natriuretic peptide (AtPNP-A). The pre-processed sequence of AtPNP-A (UniProt AC: Q9ZV52) consists of 130 residues and contains a signal peptide in its N-terminal (residues 1–25), responsible for its secretion. The active domain of the AtPNP-A molecule (AtPNP-A36–69) corresponds to the 36–69 region (green box).
In the early 2000s, Ludidi et al. suggested that the hANP displays a distant sequence homology (ca. 30%) with the 34aa active domain of AtPNP-A and presented their alignment, based on their sequence similarity[20][29]. The two proteins have a similar course of action after secretion, as they bind to membrane receptors and activate similar signaling pathways involved in ion balance and water movement regulation [13][22]. Additionally, it is known that all vertebrate NPs are conserved and share a common structure of a 17aa ring formed by a disulfide bond, that has been proved to be essential for their biological activity [30]. Experiments that followed, showed that animal NPs can induce responses in plants similar to PNPs and vice versa [31][32], while Gehring et al. provided evidence that the NPs must maintain their 17aa loop structure in order to exhibit this biological activity in plants [33]. Especially in the case of hANP and AtPNP-A, it has been shown that they induce similar functional effects in plants, while AtPNP-A can affect cardiomyoblast cell lines, inducing apoptosis similarly to hANP [32][34][35][36]. When it comes to other PNPs, multiple alignments of all the recently available PNPs from several plant species show relative conservation in the sequences that correspond to the 34aa domain of AtPNP-A. It is noteworthy that all the PNP sequences include a pair of cysteine residues, a fact which indicates that the loop ring structure is presumably present and essential for the biological activity of PNPs.
Taking into consideration the well-established amyloidogenicity of hANP [11][37], along with the functional similarity to its plant homologue, we decided to investigate the aggregation properties of the homologous region that corresponds to the biologically active and conserved functional domain of AtPNP-A [32][33]. Using biophysical methods, we demonstrate that this region (amino acid residues 36–69) of AtPNP-A self-assembles into fibrils with characteristic amyloid properties in three different pH conditions. We also provide computational evidence for the potential implication of A.thaliana proteins—associated with AtPNP-A—in amyloid fibril formation. Our data add to the heterogeneous list of proteins shown to form amyloids in plants and is expected to promote other computational and experimental studies on plants in the field of amyloid biology.
2. Implication of Different PH Values on AtPNP-A36–69 Fibril Formation
The results suggest that plant amyloid fibrils have similar properties to the ones of mammalian origin, since in many cases the fibrillar assemblies of mammalian proteins differ depending on the incubation conditions, such as precursor monomer concentration [38] and pH [39][40][41]. Specifically, when it comes to pH changes, a-helix to β-strand conversion occurs in a variety of amyloid-forming proteins, such as Aβ peptide [42], β2-microglobulin [43], and transthyretin [44]. In most of these cases, amyloid formation is enhanced at slightly acidic pH values, while in basic pH values, these proteins form amorphous aggregates or a small amount of amyloids [45][46]. However, this work shows that AtPNP-A36–69 tends to form more robust and well-defined amyloid fibrils at higher pH values, than those at lower pH values, highlighting a difference between mammalian and plant proteins. AtPNP-A is a mobile molecule, mainly expressed under abiotic and biotic stress conditions [20], which favor the increase of pH of the apoplastic space of cells, as well as that of xylem sap [47][48]. These facts may imply a functional role of AtPNP-A when in an amyloid state, under stress and increasing pH values.
PNPs have a significant biotechnological interest regarding the genetic improvement of plants towards drought stress conditions and pathogen [22][49][50]. The AtPNP-A36–69 peptide constitutes the active domain throughout the PNP family and is a target for the genetic modification of the plant. The amyloidogenic properties of this sequence should be taken into consideration whenever we seek to overexpress this peptide [21].
3. Analysis of the AtPNP-A Network
Research interest has only recently started to focus on plant proteins with amyloidogenic properties. Computational analysis predicted that the Arabidopsis thaliana proteome appears to be abundant in proteins with amyloidogenic regions and a variety of different proteins were predicted as amyloidogenic candidates [51]. In this study, we wanted to find out if other proteins of Arabidopsis thaliana have been studied experimentally regarding their amyloidogenic properties, as well as their association with AtPNP-A, in order to unravel the importance of its ability to form amyloid fibrils. For this reason, we constructed the AtPNP-A network (Figure 2). From the 32 proteins of the network, only the pathogenesis-related protein-1 (PR1) has been studied regarding its amyloidogenic properties, showing that plant proteins are understudied regarding their amyloidogenic potential. Olrichs et al. tested the amyloidogenic potential of PR-1 under conditions which promote the amyloid formation of its respective mammalian counterpart [52]. As the authors point out, in the plant extracellular environment, PR-1 interacts with a completely different range of biomolecules or pathogens, and thus it is expected to not form amyloid fibrils under mammalian micro-environmental conditions, even though it contains “aggregation-prone” regions [52]. Our study of AtPNP-A produced similar results, supporting the conclusions of their study.
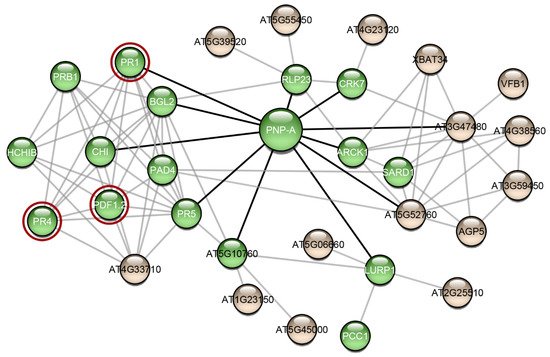
Figure 2. The interaction network of AtPNP-A. Interaction data for the creation of this network were gathered from the publicly available database STRING [53]. The network was visualized using Cytoscape 3.7.2 [54], a freely available platform for biological network visualization and analysis. The network consists of 32 nodes and 85 edges. Proteins are depicted as nodes and interactions as edges. Three of the proteins of the network (red circles) are associated with amyloid-forming proteins. Green-colored nodes are proteins that functional enrichment analysis indicated their association with defense responses. Black-colored edges are the interactions of AtPNP-A and grey-colored edges are the interactions of the AtPNP-A first neighbors.
Another interesting observation that emerged from the study of the AtPNP-A network, is that most of the proteins in this network have been associated with functions related to defense responses (Figure 2—green spheres). Computational analysis showed that proteins with predicted “aggregation-prone” regions participate in defense responses, in the majority of plant species, implying that amyloid formation has a part in their function [51]. In fact, several plant defense proteins have been shown to have amyloid-like properties in vivo and in vitro. Vicilin, a seed storage protein, has been demonstrated to form amyloid fibrils both in vivo and in vitro. In particular, Antonets et al. showed that vicilin amyloids are present in pea seeds and its amyloid accumulation increased during seed maturation. Additionally, vicilin amyloids are toxic for fungi and bacteria, suggesting a functional dualism, being both a storage and a pathogen-defense protein [55]. Interestingly, vicilin consists of two β-barrel domains, cupin-1.1 and cupin-1.2, which also form amyloid fibrils in vitro. A homology model illustrating the native fold of AtPNP-A, as well as the model of the native state of Xanthomonas axonopodis PNP-like molecule, revealed that it may adopt a double-psi β-barrel structure, which is comprised of 6 β-strands, 2 α-helices, and 2 protruding psi loops [56]. According to this model, the active domain of AtPNP-A consists of two β-strands connected via an α-helix, which is in agreement with the results of the secondary structure prediction tool PORTER [57]. Moreover, taking a closer look at the secondary structure prediction of SECSTR [58], AtPNP-A has regions with a propensity to form both α-helices and β-strands. Such sequences are characterized as “chameleon” sequences and, intrinsically, tend to alter their conformation depending on the environment [59]. Further studies regarding the native structure of AtPNP-A are needed to gain insight into its different roles and in order to unravel whether its β-barrel structure plays a role to the formation of amyloid.
RsAFP-19, a segment of the Raphanus sativus antifungal protein 1 and 2 [60], Cn-AMP2 of Cocos nucifera [61], and pro-hevein from Hevea brasiliensis [62], all have antimicrobial or antifungal properties while being amyloidogenic. Regarding the AtPNP-A network proteins, defensin-like protein 16 (Figure 2—PDF1.2) has high sequence similarity with the Raphanus sativus antifungal protein 1 (96%) and 2 (90%), suggesting that PDF1.2 likely possesses the same amyloidogenic properties as these proteins. Additionally, hevein-like pre-pro-protein (Figure 2—PR4), which is also found in the AtPNP-A network, has approximately 50% identity with pro-hevein. Therefore, it is possible that the respective A. thaliana defense proteins also form amyloid fibrils, a property which may be related to their function.
Specifically, it is believed that AtPNP-A is implicated in defense mechanisms as well. It has been shown that the transcriptional activation of AtPNP-A is triggered after a pathogen invasion, such as Agrobacterium tumefaciens [50]. Moreover, AtPNP-A is co-expressed with the Systemic Acquired Resistance marker genes—genes associated with defense against pathogens—and triggers the expression of many other genes important for plant defense [50][63]. These observations led Meier et al. to suggest the classification of AtPNP-A as a PR protein [50]. Consequently, the amyloidogenic properties of AtPNP-A may play a central role in the critical defense response mechanism of the plant, which remains to be uncovered.
This entry is adapted from the peer-reviewed paper 10.3390/plants11010009
References
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890.
- Chiti, F.; Dobson, C.M. Amyloid formation by globular proteins under native conditions. Nat. Chem. Biol. 2009, 5, 15–22.
- Cliffe, R.; Sang, J.C.; Kundel, F.; Finley, D.; Klenerman, D.; Ye, Y. Filamentous Aggregates Are Fragmented by the Proteasome Holoenzyme. Cell Rep. 2019, 26, 2140–2149.e3.
- Dobson, C.M.; Knowles, T.P.J.; Vendruscolo, M. The Amyloid Phenomenon and Its Significance in Biology and Medicine. Cold Spring Harb. Perspect. Biol. 2020, 12, a033878.
- Nastou, K.C.; Nasi, G.I.; Tsiolaki, P.L.; Litou, Z.I.; Iconomidou, V.A. AmyCo: The amyloidoses collection. Amyloid Int. J. Exp. Clin. Investig. 2019, 26, 112–117.
- Iconomidou, V.A.; Vriend, G.; Hamodrakas, S.J. Amyloids protect the silkmoth oocyte and embryo. FEBS Lett. 2000, 479, 141–145.
- Iconomidou, V.A.; Hamodrakas, S.J. Natural protective amyloids. Curr. Protein Pept. Sci. 2008, 9, 291–309.
- Johansson, B.; Westermark, P. The relation of atrial natriuretic factor to isolated atrial amyloid. Exp. Mol. Pathol. 1990, 52, 266–278.
- Gardner, D.G. Molecular biology of the natriuretic peptides. Trends Cardiovasc. Med. 1994, 4, 159–165.
- Pucci, A.; Wharton, J.; Arbustini, E.; Grasso, M.; Diegoli, M.; Needleman, P.; Vigano, M.; Polak, J.M. Atrial amyloid deposits in the failing human heart display both atrial and brain natriuretic peptide-like immunoreactivity. J. Pathol. 1991, 165, 235–241.
- Linke, R.P.; Voigt, C.; Störkel, F.S.; Eulitz, M. N-terminal amino acid sequence analysis indicates that isolated atrial amyloid is derived from atrial natriuretic peptide. Virchows Archiv. B Cell Pathol. Incl. Mol. Pathol. 1988, 55, 125–127.
- Ballermann, B.J.; Bloch, K.D.; Seidman, J.G.; Brenner, B.M. Atrial natriuretic peptide transcription, secretion, and glomerular receptor activity during mineralocorticoid escape in the rat. J. Clin. Investig. 1986, 78, 840–843.
- Baxter, J.D.; Lewicki, J.A.; Gardner, D.G. Atrial Natriuretic Peptide—Review. Nat. Biotechnol. 1988, 6, 529–546.
- Vesely, D.L.; Giordano, A.T. Atrial natriuretic peptide hormonal system in plants. Biochem. Biophys. Res. Commun. 1991, 179, 695–700.
- Vesely, D.L.; Gower, W.R., Jr.; Giordano, A.T. Atrial natriuretic peptides are present throughout the plant kingdom and enhance solute flow in plants. Am. J. Physiol. 1993, 265, E465–E477.
- Billington, T.; Pharmawati, M.; Gehring, C.A. Isolation and immunoaffinity purification of biologically active plant natriuretic peptide. Biochem. Biophys. Res. Commun. 1997, 235, 722–725.
- Pharmawati, M.; Gehring, C.A.; Irving, H.R. An immunoaffinity purified plant natriuretic peptide analogue modulates cGMP levels in the Zea mays root stele. Plant Sci. 1998, 137, 107–115.
- Maryani, M.M.; Shabala, S.N.; Gehring, C.A. Plant natriuretic peptide immunoreactants modulate plasma-membrane H(+) gradients in Solanum tuberosum L. leaf tissue vesicles. Arch. Biochem. Biophys. 2000, 376, 456–458.
- Maryani, M.M.; Bradley, G.; Cahill, D.M.; Gehring, C.A. Natriuretic peptides and immunoreactants modify osmoticum-dependent volume changes in Solanum tuberosum L. mesophyll cell protoplasts. Plant Sci. 2001, 161, 443–452.
- Morse, M.; Pironcheva, G.; Gehring, C. AtPNP-A is a systemically mobile natriuretic peptide immunoanalogue with a role in Arabidopsis thaliana cell volume regulation. FEBS Lett. 2004, 556, 99–103.
- Wang, Y.H.; Gehring, C.; Cahill, D.M.; Irving, H.R. Plant natriuretic peptide active site determination and effects on cGMP and cell volume regulation. Funct. Plant Biol. 2007, 34, 645–653.
- Meier, S.; Irving, H.; Gehring, C. Plant natriuretic peptides—Emerging roles in fluid and salt balance. In Cardiac Hormones; Vesely, D.L., Ed.; Transworld Research Network: Kerala, India, 2008.
- Lindsey, K.; Casson, S.; Chilley, P. Peptides: New signalling molecules in plants. Trends Plant Sci. 2002, 7, 78–83.
- Kwezi, L.; Meier, S.; Mungur, L.; Ruzvidzo, O.; Irving, H.; Gehring, C. The Arabidopsis thaliana brassinosteroid receptor (AtBRI1) contains a domain that functions as a guanylyl cyclase in vitro. PLoS ONE 2007, 2, e449.
- Turek, I.; Gehring, C. The plant natriuretic peptide receptor is a guanylyl cyclase and enables cGMP-dependent signaling. Plant Mol. Biol. 2016, 91, 275–286.
- Rafudeen, S.; Gxabab, G.; Makgokea, G.; Bradley, G.; Pironcheva, G.; Raitt, L.; Irving, H.; Gehring, C. A role for plant natriuretic peptide immuno-analogues in NaCl− and drought-stress responses. Physiol. Plant. 2003, 119, 554–562.
- Ruzvidzo, O.; Donaldson, L.; Valentine, A.; Gehring, C. The Arabidopsis thaliana natriuretic peptide AtPNP-A is a systemic regulator of leaf dark respiration and signals via the phloem. J. Plant Physiol. 2011, 168, 1710–1714.
- Wang, Y.H.; Gehring, C.; Irving, H.R. Plant natriuretic peptides are apoplastic and paracrine stress response molecules. Plant Cell Physiol. 2011, 52, 837–850.
- Ludidi, N.N.; Heazlewood, J.L.; Seoighe, C.; Irving, H.R.; Gehring, C.A. Expansin-like molecules: Novel functions derived from common domains. J. Mol. Evol. 2002, 54, 587–594.
- Yandle, T.G. Biochemistry of natriuretic peptides. J. Int. Med. 1994, 235, 561–576.
- Gehring, C.A.; Khalid, K.M.; Toop, T.; Donald, J.A. Rat natriuretic peptide binds specifically to plant membranes and induces stomatal opening. Biochem. Biophys. Res. Commun. 1996, 228, 739–744.
- Gehring, C.; Irving, H. Plant natriuretic peptides: Systemic regulators of plant homeostasis and defense that can affect cardiomyoblasts. J. Investig. Med. Off. Publ. Am. Fed. Clin. Res. 2013, 61, 823–826.
- Gehring, C.A. Natriuretic Peptides—A New Class of Plant Hormone? Ann. Bot. 1999, 83, 329–334.
- Wang, Y.H.; Ahmar, H.; Irving, H.R. Induction of apoptosis by plant natriuretic peptides in rat cardiomyoblasts. Peptides 2010, 31, 1213–1218.
- Wu, C.F.; Bishopric, N.H.; Pratt, R.E. Atrial natriuretic peptide induces apoptosis in neonatal rat cardiac myocytes. J. Biol. Chem. 1997, 272, 14860–14866.
- Pharmawati, M.; Maryani, M.; Nikolakopoulos, T.; Gehring, C.; Irving, H. Cyclic GMP modulates stomatal opening induced by natriuretic peptides and immunoreactive analogues. Plant Physiol. Biochem. 2001, 39, 385–394.
- Louros, N.N.; Iconomidou, V.A.; Tsiolaki, P.L.; Chrysina, E.D.; Baltatzis, G.E.; Patsouris, E.S.; Hamodrakas, S.J. An N-terminal pro-atrial natriuretic peptide (NT-proANP) ‘aggregation-prone’ segment involved in isolated atrial amyloidosis. FEBS Lett. 2014, 588, 52–57.
- Bauer, H.H.; Aebi, U.; Haner, M.; Hermann, R.; Muller, M.; Merkle, H.P. Architecture and polymorphism of fibrillar supramolecular assemblies produced by in vitro aggregation of human calcitonin. J. Struct. Biol. 1995, 115, 1–15.
- Sneideris, T.; Darguzis, D.; Botyriute, A.; Grigaliunas, M.; Winter, R.; Smirnovas, V. pH-Driven Polymorphism of Insulin Amyloid-Like Fibrils. PLoS ONE 2015, 10, e0136602.
- Iannuzzi, C.; Borriello, M.; Portaccio, M.; Irace, G.; Sirangelo, I. Insights into Insulin Fibril Assembly at Physiological and Acidic pH and Related Amyloid Intrinsic Fluorescence. Int. J. Mol. Sci. 2017, 18, 2551.
- Goldsbury, C.S.; Cooper, G.J.; Goldie, K.N.; Muller, S.A.; Saafi, E.L.; Gruijters, W.T.; Misur, M.P.; Engel, A.; Aebi, U.; Kistler, J. Polymorphic fibrillar assembly of human amylin. J. Struct. Biol. 1997, 119, 17–27.
- Barrow, C.J.; Yasuda, A.; Kenny, P.T.; Zagorski, M.G. Solution conformations and aggregational properties of synthetic amyloid beta-peptides of Alzheimer’s disease. Analysis of circular dichroism spectra. J. Mol. Biol. 1992, 225, 1075–1093.
- McParland, V.J.; Kad, N.M.; Kalverda, A.P.; Brown, A.; Kirwin-Jones, P.; Hunter, M.G.; Sunde, M.; Radford, S.E. Partially unfolded states of beta(2)-microglobulin and amyloid formation in vitro. Biochemistry 2000, 39, 8735–8746.
- Lai, Z.; Colon, W.; Kelly, J.W. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry 1996, 35, 6470–6482.
- Srinivasan, R.; Jones, E.M.; Liu, K.; Ghiso, J.; Marchant, R.E.; Zagorski, M.G. pH-dependent amyloid and protofibril formation by the ABri peptide of familial British dementia. J. Mol. Biol. 2003, 333, 1003–1023.
- Xu, W.; Zhang, C.; Derreumaux, P.; Graslund, A.; Morozova-Roche, L.; Mu, Y. Intrinsic determinants of Abeta(12-24) pH-dependent self-assembly revealed by combined computational and experimental studies. PLoS ONE 2011, 6, e24329.
- Malcheska, F.; Ahmad, A.; Batool, S.; Muller, H.M.; Ludwig-Muller, J.; Kreuzwieser, J.; Randewig, D.; Hansch, R.; Mendel, R.R.; Hell, R.; et al. Drought-Enhanced Xylem Sap Sulfate Closes Stomata by Affecting ALMT12 and Guard Cell ABA Synthesis. Plant Physiol. 2017, 174, 798–814.
- Geilfus, C.M. The pH of the Apoplast: Dynamic Factor with Functional Impact under Stress. Mol. Plant 2017, 10, 1371–1386.
- Meier, S.; Madeo, L.; Ederli, L.; Donaldson, L.; Pasqualini, S.; Gehring, C. Deciphering cGMP signatures and cGMP-dependent pathways in plant defence. Plant Signal. Behav. 2009, 4, 307–309.
- Meier, S.; Bastian, R.; Donaldson, L.; Murray, S.; Bajic, V.; Gehring, C. Co-expression and promoter content analyses assign a role in biotic and abiotic stress responses to plant natriuretic peptides. BMC Plant Biol. 2008, 8, 24.
- Antonets, K.S.; Nizhnikov, A.A. Predicting Amyloidogenic Proteins in the Proteomes of Plants. Int. J. Mol. Sci. 2017, 18, 2155.
- Olrichs, N.K.; Mahalka, A.K.; Kaloyanova, D.; Kinnunen, P.K.; Bernd Helms, J. Golgi-Associated plant Pathogenesis Related protein 1 (GAPR-1) forms amyloid-like fibrils by interaction with acidic phospholipids and inhibits Abeta aggregation. Amyloid Int. J. Exp. Clin. Investig. 2014, 21, 88–96.
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613.
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504.
- Antonets, K.S.; Belousov, M.V.; Sulatskaya, A.I.; Belousova, M.E.; Kosolapova, A.O.; Sulatsky, M.I.; Andreeva, E.A.; Zykin, P.A.; Malovichko, Y.V.; Shtark, O.Y.; et al. Accumulation of storage proteins in plant seeds is mediated by amyloid formation. PLoS Biol. 2020, 18, e3000564.
- Nembaware, V.; Seoighe, C.; Sayed, M.; Gehring, C. A plant natriuretic peptide-like gene in the bacterial pathogen Xanthomonas axonopodis may induce hyper-hydration in the plant host: A hypothesis of molecular mimicry. BMC Evol. Biol. 2004, 4, 10.
- Mirabello, C.; Pollastri, G. Porter, PaleAle 4.0: High-accuracy prediction of protein secondary structure and relative solvent accessibility. Bioinformatics 2013, 29, 2056–2058.
- Hamodrakas, S.J. A protein secondary structure prediction scheme for the IBM PC and compatibles. Bioinformatics 1988, 4, 473–477.
- Kabsch, W.; Sander, C. On the use of sequence homologies to predict protein structure: Identical pentapeptides can have completely different conformations. Proc. Natl. Acad. Sci. USA 1984, 81, 1075–1078.
- Garvey, M.; Meehan, S.; Gras, S.L.; Schirra, H.J.; Craik, D.J.; Van der Weerden, N.L.; Anderson, M.A.; Gerrard, J.A.; Carver, J.A. A radish seed antifungal peptide with a high amyloid fibril-forming propensity. Biochim. Biophys. Acta 2013, 1834, 1615–1623.
- Gour, S.; Kaushik, V.; Kumar, V.; Bhat, P.; Yadav, S.C.; Yadav, J.K. Antimicrobial peptide (Cn-AMP2) from liquid endosperm of Cocos nucifera forms amyloid-like fibrillar structure. J. Pept. Sci. Off. Publ. Eur. Pept. Soc. 2016, 22, 201–207.
- Berthelot, K.; Lecomte, S.; Coulary-Salin, B.; Bentaleb, A.; Peruch, F. Hevea brasiliensis prohevein possesses a conserved C-terminal domain with amyloid-like properties in vitro. Biochim. Biophys. Acta 2016, 1864, 388–399.
- Turek, I.; Wheeler, J.I.; Gehring, C.; Irving, H.R.; Marondedze, C. Quantitative proteome changes in Arabidopsis thaliana suspension-cultured cells in response to plant natriuretic peptides. Data Brief 2015, 4, 336–343.
This entry is offline, you can click here to edit this entry!
