High-resolution episcopic microscopy (HREM) is a three-dimensional (3D) episcopic imaging modality based on the acquisition of two-dimensional (2D) images from the cut surface of a block of tissue embedded in resin. Such images, acquired serially through the entire length/depth of the tissue block, are aligned and stacked for 3D reconstruction. HREM has proven to be specifically advantageous when integrated in correlative multimodal imaging (CMI) pipelines. CMI creates a composite and zoomable view of exactly the same specimen and region of interest by (sequentially) correlating two or more modalities. CMI combines complementary modalities to gain holistic structural, functional, and chemical information of the entire sample and place molecular details into their overall spatiotemporal multiscale context. HREM has an advantage over in vivo 3D imaging techniques on account of better histomorphologic resolution while simultaneously providing volume data. HREM also has certain advantages over ex vivo light microscopy modalities. The latter can provide better cellular resolution but usually covers a limited area or volume of tissue, with limited 3D structural context.
1. Introduction
High-resolution episcopic microscopy (HREM) is a 3D episcopic imaging modality that is capable of generating volume data from whole embryos and isolated tissue specimens (biopsies). HREM is essentially an innovative adaptation of routine histology and light microscopy techniques which have been described in detail previously [
1] (pp. I.2.g-1–I.2.g-9) [
2,
3]. As in routine histology, fixed samples are dehydrated in increasing concentrations of ethanol or methanol. Then, samples are infiltrated with and embedded in plastic methacrylate resin, which is coloured with eosin B. The polymerised resin blocks are then physically sectioned. After each section, an image is captured directly from the freshly exposed block surface using a fluorescence microscope equipped with a GFP or YFP filter-set. The preceding eosin staining provides unspecific contrast of the embedded tissues and ensures that only structures on the surface of the block are visualised. Both the optics set-up and the sample retain the same position during this procedure, and thus the images can be reconstructed to obtain three-dimensional data of the sample [
1] (pp. I.2.g-1–I.2.g-9) [
3,
4,
5,
6].
Correlated multimodal imaging (CMI) represents a combinatorial approach of multiple sequential or parallel in vivo and ex vivo imaging and analysing modalities on the same tissue specimen. CMI is capable of providing structural and functional information on a tissue sample that is visualised at different lateral resolutions and penetration depths across relevant scales. This essentially involves the application of two or more complementary modalities, which in combination, provide a more informative and composite view of normal and abnormal features of the tissue specimen [
7,
8,
9]. Prominent examples of CMI approaches that combine two modalities include:
-
Combinations of radiology (including computed tomography (CT) and magnetic resonance imaging (MRI)) and pathology (including histopathology) provide valuable clinical diagnostic and preclinical information for better patient care or further biomedical discovery [
10,
11].
-
Combinations of light microscopy and electron microscopy (EM) have been in vogue for diagnostic and research applications. In clinical and some preclinical settings, such combinations are well established in the forms of histopathology (HP) and ultrastructural pathology. In recent times, the combined application of HP, immunohistochemistry, and electron microscopy (scanning and transmission) is central to the characterisation of pulmonary lesions in fatal cases of COVID-19 [
12]. In biological research, the combination of these two microscopic modalities is designated as correlative light and electron microscopy (CLEM) [
13,
14].
More recently, there have been CMI efforts to combine micro-magnetic resonance imaging (micro-MRI), micro-computed tomography (micro-CT), micro-Positron emission tomography (microPET), HREM, and HP, to evaluate murine tumour vasculature [
15] and murine non-neoplastic vascular lesions [
16] across scales. These efforts highlighted the benefits and challenges of such multimodal efforts. As expected, micro-MRI and micro-CT provided good in vivo anatomic resolution with lower tissue sensitivity, whereas micro-PET was highly sensitive but had poor spatial resolution. HREM and HP achieved the highest spatial histomorphologic resolution, but required irreversible ex vivo processing (trimming, embedding, and sectioning) [
1] (pp. I.2.g-1–I.2.g-9, I.2.i-1–I.2.i-13) [
15,
16]. Despite some of the disadvantages, the combination of in vivo imaging followed by ex vivo processing for sequential or parallel HREM and HP can provide distinct advantages in the characterisation of sub-macroscopic and micro-anatomic morphologic defects [
1] (pp. I.2.g-1–I.2.g-9, I.2.i-1–I.2.i-13) [
15,
16].
Due to the plethora of potentially beneficial imaging combinations, CMI has been used to tackle a variety of research questions and will continue to broaden the accessible biomedical information significantly. In this review, we highlight HREM as versatile imaging technology that helps bridge preclinical and biological imaging and integrate in vivo dynamics with ex vivo high resolution in a variety of model organisms.
Advantages and Limitations of HREM in a Multimodal Context
The strengths and limitations of HREM are represented in
Table 1 in the context of other imaging modalities that, to our knowledge, HREM has been combined with. As can be seen in
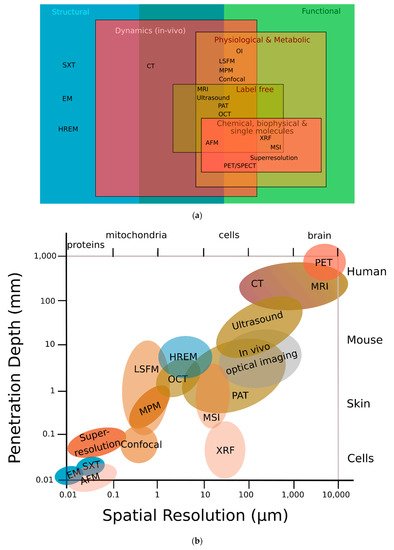
Figure 1a,b, HREM assesses structural information, occupies a resolution niche that is not accessible to any other imaging modality, and connects preclinical imaging (such as micro-MRI, micro-CT, or small-animal ultrasound (US)) with biological microscopy (such as advanced fluorescence, electron, or atomic force microscopy) [
7].
Figure 1. HREM can provide both high image resolution and high sample penetration depths. (
a) Classification of different modalities according to their function; HREM offers structural information that most other modalities cannot. (
b) HREM lies in the middle-field concerning both penetration depth and spatial resolution. Adapted with permission from [
13]. (AFM) atomic force microscopy; (CT) computed tomography; (EM) electron microscopy; (HREM) high-resolution episcopic microscopy; (LSFM) light sheet fluorescence microscopy; (MPM) multiphoton microscopy; (MRI) magnetic resonance imaging; (MSI) mass spectrometry imaging; (OCT) optical coherence tomography; (OI) optical interferometry; (PAT) photoacoustic tomography; (PET) positron emission tomography; (SPECT) single-photon emission computed tomography; (SXT) soft X-ray tomography; (XRF) X-ray fluorescence.
Table 1. Different modalities that have previously been combined in CMI studies in combination with HREM, and their imaging parameters, advantages, and limitations [
1,
7,
17,
18]. (CT) computed tomography; (HP) histopathology; (HREM) high-resolution episcopic microscopy; (MRI) magnetic resonance imaging; (OCT) optical coherence tomography; (PAT) photoacoustic tomography; (PET) positron emission tomography; (US) ultrasound.
| Modality |
Contrast |
Penetration (mm) |
Lateral Resolution (µm) |
VOI |
Advantages |
Limitations |
| micro-MRI |
Emitted RF signal after nuclear spin excitation |
>500 |
≤100 |
whole organism |
- -
-
non-ionising radiation
- -
-
excellent soft tissue contrast
- -
-
biochemical information (spectroscopy)
|
- -
-
expensive equipment
- -
-
high maintenance costs
|
| micro-US |
Acoustic impedance
between tissue interfaces; detection of echoes from moving particles |
<150 |
30–800 |
whole organism |
- -
-
high temporal and spatial resolution
- -
-
portable instrumentation
- -
-
cost-efficient
|
- -
-
limited tissue penetration
- -
-
poor contrast
- -
-
difficult to quantitate
|
| PAT |
Acoustic waves generated by optical absorption of tissue chromophores |
~10 |
~40 |
10 × 10 mm2 |
- -
-
in vivo
- -
-
penetration depth
- -
-
endogenous and exogenous contrast
|
- -
-
resolution
- -
-
speed
- -
-
structural contrast
|
| OCT |
Optical scattering based on refractive index changes;
motion contrast due to blood flow (OCT angiography) |
~1–2 |
1–10 (diffraction limited) |
10 × 10 mm2 |
- -
-
in vivo
- -
-
fast
- -
-
non-invasive
- -
-
label-free
- -
-
morphology
- -
-
quantitative blood flow
|
- -
-
limited molecular information
- -
-
reduced sub-cellular resolution
- -
-
minimum blood flow required
|
| micro-PET |
Photon emission after positron annihilation |
>500 |
1000–2000 |
whole organism |
- -
-
high sensitivity
- -
-
fully quantitative
- -
-
broad range of applications (imaging agent dependent)
- -
-
dynamic measurements
|
- -
-
use of radioactive agents
- -
-
highly specialised equipment and staff required
- -
-
high costs
|
| micro-CT |
Differential X-ray attenuation of tissues related to their density |
>500 |
≤100 |
whole organism |
- -
-
excellent bone imaging
|
- -
-
radiation dose
- -
-
low soft-tissue contrast (use of contrast agents)
|
| HREM |
Light scattering based on unspecific eosin staining |
Sample size up to 12 mm in thickness |
>1 |
8 mm × 8 mm × 12 mm |
- -
-
digital volumes in histologic quality at high resolution
|
- -
-
whole-mount contrasting of specimens
- -
-
time-consuming (fixation and acquisition of 3D volume)
- -
-
ex vivo, no dynamics, structural data only
|
| HP |
Light scattering
(various staining methods impart colour and contrast to cellular and tissue components) |
<0.1 |
>500 |
up to 1 mm3 |
- -
-
evaluation of overall tissue features at low costs
- -
-
excellent cellular detail at light microscopic resolution
- -
-
spatial contextual correlation of microscopic morphology
|
- -
-
2D and static
- -
-
detailed evaluation (especially of abnormal features in lesions) requires additional expertise
|
In imaging, penetration depth comes at the expense of lateral resolution, which restricts the scope of 3D imaging of small animals at micrometre resolution. HREM covers the mesoscopic imaging range, which refers to techniques that allow the 3D visualisation of large samples at the millimetre to centimetre scale. HREM allows the combination of a large field of view and the ability to image thick tissues of several millimetres in thickness with a high-micrometre resolution. Thus, fine structures can be analysed in the context of the overall morphology of surrounding tissues or even whole organisms. The large penetration depth of HREM is achieved by physical sectioning, which, as a downside, limits its application to dead, sacrificed samples. Since the probed volume is sectioned, images are captured from the block surface and the whole 3D sample is reconstructed virtually. There is no need for clearing the sample, as, for example, is the case in light sheet microscopy, which is best suited to optically homogeneous and relatively transparent samples. In addition, while light sheet microscopy can achieve higher resolution than HREM for similar probed volumes, the higher resolution in light sheet microscopy comes with a narrower field of view that would need to be tiled across large specimens.
2. State-of-the-Art
HREM is compatible with a wide range of fixatives and various contrast agents, and tissue processing for HREM does not require special chemicals except for resin embedding and contrasting with eosin. Therefore, HREM can be easily combined with almost all upstream and downstream techniques in multimodal imaging pipelines. It proved to be the method of choice to correlatively reconstruct tumour capillaries and murine vasculature of sufficient contrast and quality at micrometre resolution in selected volumes of interest (VOIs), and to visualise minute anatomical structures and volume displays of mouse, chick, and zebrafish embryos (such as heart malformations) in combination with optical coherence tomography (OCT), photoacoustic tomography (PAT), micro-US, micro-CT, and micro-MRI. HREM was shown to be compatible with contrast agent-enhanced CT and micro-MRI and with the fixation, staining, and dehydration media used after tumour and embryo removal for ex vivo micro-CT or HP. Even though HREM is destructive to the tissue, it can be combined and correlated with HP by collecting physical sections for subsequent examination.
2.1. HP and HREM: Mutual Complementarity
HP is well established in several non-clinical settings including translational biomedical research, model organism phenotyping, preclinical therapeutic discovery, and preclinical safety assessment. In these settings, HP is integral to study specific data generation and validation [
1] (pp. I.2.i-1–I.2.i-13) [
19,
20,
21,
22,
23]. HREM has been most impactful in the phenotyping of embryos and characterisation of structural developmental anomalies. HREM has also found some application in the three-dimensional visualisation of structures in tissues such as human skin and human liver, and in the characterisation of aberrant tumour vasculature and vessel wall lesions in mouse models [
1] (pp. I.2.g-1–I.2.g-9) [
15,
16]. HREM is not typically featured in routine diagnostics, therapeutic discovery, or safety assessment [
1] (pp. I.2.i-1–I.2.i-13) [
19,
20,
21,
22,
23].
Founded on the same principles of histology, HREM and HP are modalities that generate valuable morphologic data at relatively low costs [
1] (pp. I.2.g-1–I.2.g-9, I.2.i-1–I.2.i-13) [
6,
19,
21,
22].
HP is based on the evaluation of tissue sections on glass slides, typically by expert pathologists, followed by the acquisition of 2D images at different magnifications to represent specific features of interest observed during evaluation. HREM on the other hand is based on the acquisition of images from the cut surface of the block (rather than the section itself) through the entire thickness of the embedded tissue. The use of the lower magnification objectives incorporates a greater area of the tissue for serial imaging and 3D reconstruction but provides lower cellular detail than available in HP images.
HREM and HP focus on imaging different surfaces of the embedded tissue (block surface versus tissue section) while following the same fundamental principles of histology and light microscopy. They are closely related and mutually complementary modalities, which—when properly combined—can provide histomorphologic information with better 2D cellular detail and 3D spatial context [
1] (pp. I.2.g-1–I.2.g-9) [
3,
4,
5,
6,
15,
16]. The similarities and differences between certain aspects of the two modalities are summarised in
Table 2.
Table 2. Comparison of selected aspects of HP and HREM.
| |
HP 1 |
HREM 2 |
| Fixation |
Predominantly aldehyde-based fixatives
(10% formaldehyde,
4% paraformaldehyde, Bouin’s fluid) |
Predominantly aldehyde-based fixatives
(10% formaldehyde,
4% paraformaldehyde, Bouin’s fluid) |
Processing
(infiltration) |
Automated or manual processing
Paraffin (most common)
Resin, agar, gelatine, celloidin (alternative, less common) |
Manual processing
Resin (JB-4) |
| Embedding |
Paraffin
Resin |
Resin (JB-4) |
| Sectioning |
Manual rotary microtomy
Automated microtomy |
Automated microtomy |
| Sectioning |
Single or multiple sections at specific planes of the embedded tissue for most routine diagnostic cases and discovery projects; serial sections are reserved for specialised analyses |
Serial sections (at specific intervals through the entire thickness of the block) |
| Section thickness |
1 µm to 5 µm |
1 µm to 3 µm |
| Staining |
Tissue sections are placed on glass slides and then stained |
Tissues are stained during infiltration (prior to embedding) |
| Stains |
Several histochemical stains including Hematoxylin and Eosin (H&E), Periodic Acid Schiff (PAS) and Luxol Fast Blue (LFB) |
Eosin |
| Imaging |
Light microscopy, multiple objectives and magnifications |
Light microscopy, single objective and magnification (selected at the start of sectioning) |
| Imaging surface |
Tissue section on glass slide |
Cut surface of resin block |
| Visualisation and resolution |
2D; higher histomorphologic and cellular resolution with better discernment of specific lesions 3 |
2D and (virtual/reconstructed) 3D; broader spatial resolution and architectural overview with lower cellular resolution (than HP) 3 |
| Spatial contextual analysis of molecular (protein and nucleic acid) markers |
More options for immunostaining and in situ hybridisation on paraffin embedded sections |
Fewer options for immunostaining and in situ hybridisation on JB-4 resin embedded sections |
2.2. Multimodality HREM to Image Mouse, Chick, Quail, Frog, and Zebrafish Embryos
Developmental pathology is a well-established discipline in hospital and non-hospital practice and HP is integral to diagnosing/characterising many developmental anomalies in patients and model organisms [
22,
23,
24,
25]. Congenital defects in the embryo or foetus caused by exposure to various toxic agents including certain drugs are studied under developmental toxicologic pathology. Screening for such defects in the embryos of different rodent and non-rodent species constitutes an important component of the safety assessment of new drug candidates [
20]. HP has been, and continues to be, a significant modality in the characterisation and representation of stages of normal embryonic development and certain causes of embryonic mortality [
24,
25].
Episcopic 3D imaging methods, and HREM in particular, have become established in life science research as valuable and impactful modalities for 3D visualisation and characterisation of normative and defective embryo anatomy. A large-scale mouse embryo phenotyping project designated as Deciphering the Mechanisms of Developmental Disorders (DMDD,
https://dmdd.org.uk/, accessed 25 September 2021) is significantly based upon HREM methodology [
1] (pp. I.2.g-1–I.2.g-9) [
5,
6].
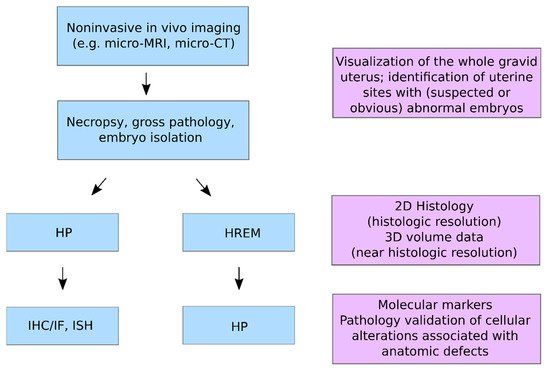
Non-invasive in vivo imaging modalities such as micro-CT and micro-MRI are excellent tools for screening developmental phenotypes and monitoring lesions as they evolve. These modalities can provide reliable information on the location, overall structure, volume, distribution, and number of lesions, but lack sufficient resolution to distinguish specific histomorphologic lesions and cellular alterations. In some contexts, ex vivo information obtained from in vivo imaging would be incorporated into downstream pathology workflows to enable more detailed HP evaluation. Such workflows are common in clinical diagnostic and non-clinical discovery settings wherein MRI, gross pathology, and HP are combined to arrive at diagnostic conclusions or to derive relevant morphologic data [
10,
11,
25,
26]. Given these considerations, a CMI workflow as illustrated in
Figure 2 would be beneficial for the systematic evaluation of embryonic/foetal anomalies in basic life science and non-clinical translational research settings.
Figure 2. CMI workflow for embryo phenotyping in vivo imaging, HREM and HP.
In the following, we provide an exhaustive overview of published CMI workflows that specifically integrated HREM to diagnose and characterise embryos or foetuses.
2.2.1. Visualising Gene Activity within Tissue
HREM is based on unspecific contrasting and provides highly detailed information of the overall morphology of various biologic specimens. Combining this detailed structural information with the exact localisation of specifically labelled structures, gene expression and gene product patterns is a highly beneficial tool to unravel basic developmental and pathologic mechanisms. Thus, attempts to detect specifically stained structures were undertaken from the very first beginnings of HREM.
Due to methodical constraints, the visualisation of specific signals has to be performed after wholemount staining prior to the embedding of the specimen, which is challenging for large samples and hard-to-penetrate tissues. Nevertheless, it has been achieved following wholemount in situ hybridisation, wholemount immuno-staining and lacZ staining utilising two of the multiband filter sets during HREM data generation [
27]. More recently, multifluorescence HREM (MF-HREM) utilising fluorescent dyes and opaque resin has been introduced [
28].
2.2.2. Combining Micro-MRI and HREM for the Analysis of Murine Embryos
The production and screening of mutant embryos in large-scale phenotyping studies is expensive and time consuming. Imaging pipelines that facilitate rapid pre-selection of samples or definition of volumes of interest at lower resolution, which is then followed by detailed validation at higher resolution, could provide an efficient alternative.
Both micro-MRI and HREM have been established as routine tools for analysing phenotypes of genetically altered mouse embryos. While micro-MRI offers the advantage of being able to simultaneously scan multiple embryos at the organ and organ-system level in one run, HREM offers superior near-histological image quality. The combination of both techniques allows for identifying embryos of interest that show an ambiguous phenotype or a phenotype hinting at additional abnormalities with micro-MRI. These embryos are then subjected to HREM for further analysis [
26].
This also allowed for a better management of the large amounts of data typically generated during acquisition. A combination of both modalities offered a continuous high-throughput imaging pipeline.
2.2.3. From In Vivo to High Resolution Using OCT, PAT, and HREM
Chick embryos are a valuable model organism both for developmental and cancer biology. They can be used to study embryonic morphogenesis and develop experimental surgical techniques. They are also especially useful when studying tumour genesis. To establish imaging pipelines combining in vivo observations and high resolution, various modalities must be combined. OCT and PAT are techniques that have previously been successfully used in vivo [
29] and in utero [
30], respectively. A pipeline integrating OCT/PAT has been established for chick embryos at several developmental stages. Following these modalities, HREM then offered high-resolution insights into regions of interest [
31].
Overall, this pipeline has the potential to facilitate the analysis and monitoring of tumour genesis, organogenesis, and the development of vasculature in embryos.
2.2.4. CMI Pipeline to Track the Genesis of Congenital Heart Malformations
An area of interest in developmental biology is the establishment of the left-right body axis and its underlying molecular mechanisms during early development. The heart as an asymmetric organ is an important model for left-right morphogenesis, and laterality defects are the cause for many complex congenital heart defects. To track embryonic development, in vivo imaging techniques—such as small-animal US and micro-CT—are of special interest to those research questions, coupled with high-resolution analysis methods that can reveal morphological abnormalities in detail.
In the study of Desgrange et al. [
32], murine embryos at different stages of development were imaged, with a special focus on the development of the heart loop. Using micro-US, embryos were visualised in vivo at E9.5, and their positions in the uterine horns of the pregnant mouse determined. This was followed by micro-CT and HREM after sacrificing the embryos at E18.5. Because of the non-invasiveness of micro-CT, it was possible to check the situs of the visceral organs within the embryo without dissection. Afterwards, organs of interest, such as the heart, were excised and prepared for visualisation with HREM. Structural abnormalities were then determined at higher resolution.
This pipeline is not restricted to analyse laterality defects and can be easily modified to focus on other organs and developmental stages.
2.3. Multimodality HREM to Assess Murine Vasculature
Cardiovascular pathology is a well-established discipline in hospital and non-hospital practice, and HP, in tandem with in vivo imaging methods, is integral to evaluating and diagnosing a diverse array of cardiac and vascular pathologies in patients and model organisms [
19,
20,
23,
33]. HP is essential for the characterisation of vascular lesions such as mural microthrombi, atherosclerotic plaques, mural microcalcifications, autoimmune or infectious vasculitis, vascular intimal, and medial proliferation/hyperplasia, vascular tumours, and abnormal vessels in tumours [
19,
23,
33,
34]. HREM has been used for non-routine investigative analyses of blood vessels and other structures in human liver samples and human and porcine skin samples. HREM facilitated 3D representations of the arterial and venous networks in skin samples in these efforts [
1] (pp. I.2.g-1–I.2.g-9). More recently, under the auspices of CMI, HREM and HP have been applied in combination to characterise vessel wall lesions and abnormal tumour vasculature in different mouse models [
15,
16]. The combination of HREM and HP facilitated the definitive identification of blood vessels and distinction of non-vascular ductal structures [
15].
Two published studies are compiled in the following.
2.3.1. Murine Tumour Vasculature
Tumour vasculature plays a major role for tumour progression and dissemination, highlighting the importance of vascular visualisation. The application of CMI pipelines enables the depiction of multiple vascular parameters of the same tumour across scales and penetration depths.
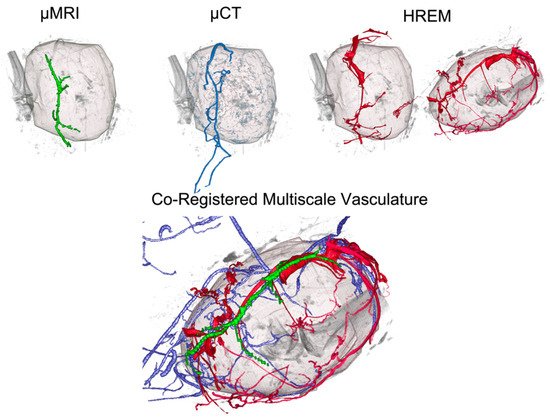
As part of a proof-of-principle study, a novel CMI pipeline was established to characterise aberrant tumour vasculature in a murine orthotopic melanoma model. This CMI approach by Zopf et al. [
15] incorporated modalities such as MRI, PET, CT, OCT, HREM, and HP (
Figure 3). This novel platform explored the feasibility of combining these technologies using an extensive image processing pipeline. In this workflow, HREM offered the highest resolution of about 3 µm. Two-dimensional HREM images offered near-histologic resolution, and the structures were further resolved and confirmed by post-HREM HP. For post-HREM HP, JB-4 resin sections were collected during HREM imaging, stained, and additionally evaluated to validate the intratumoral vessels that were segmented based on the HREM images. Correlative registration was achieved between H & E-stained JB-4 sections (collected during HREM imaging) and 2D HREM images by using the morphologic landmarks of skin, subcutaneous vessels, nerve bundles, and mammary ducts. Importantly, blood vessels did not need to be perfused or selectively contrasted for 3D imaging, which is why HREM, in addition to visualising capillaries inside and outside the tumour, also allowed one to visualise capsules, blindly terminating or beginning vessels, and short collaterals inside the tumour and necrotic tissue.
Figure 3. Integration of HREM data (shown in red) into a multimodal imaging pipeline of micro-MRI (green), micro-CT (blue), and HREM to reveal the vascular network of a murine tumour across scales. With a resolution of about 3 µm, HREM allowed the visualisation of blood vessels that were not detected by any of the other modalities. Reprinted from [
15].
2.3.2. Murine Vascular Lesions
The combination of in vivo imaging with ex vivo biological microscopy enables the visualisation of both the whole organism and single structures within. With the aim of characterising vascular lesions in a genetically engineered mouse model, a novel multimodal workflow was established as described in Keuenhof et al. [
16] by combining micro-MRI and micro-CT with HREM to identify and study vascular abnormalities using a targeted knockout mouse model (
Figure 4). The pipeline allowed the study of the same VOI for each modality at different length scales, from an overview at the organ level (micro-MRI, micro-CT) to micrometre resolution (HREM).
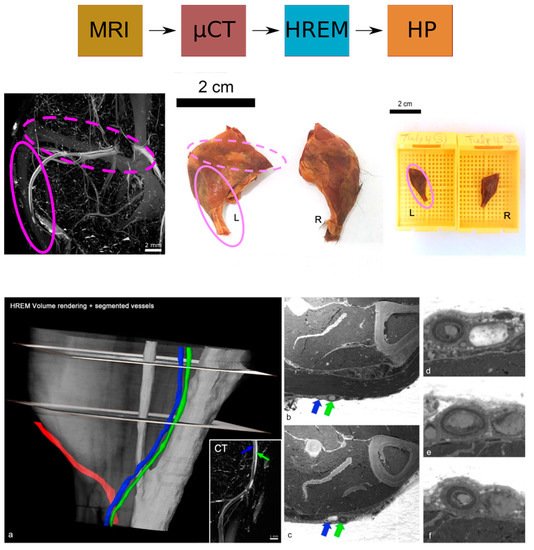
Figure 4. Identification of the VOI and segmented blood vessels to detect vascular lesions. (
a) HREM and CT (inlay) volume models displaying subcutaneous blood vessels in the colours red, blue, and green; (
b,
c) Cross-sections of the HREM stack illustrated in (
a); (
d–
f) higher magnification images of sections showing intimal hyperplasia in blood vessels. The green and blue arrow indicate the respective vessel from panel (
a). Reprinted with permission from [
16].
For an overview at the macroscopic scale of the entire mouse vasculature and the detection of suspected vascular lesions, micro-MRI was used with a lateral resolution of about 100 × 50 µm
2. With contrast-enhanced ex vivo micro-CT, blood vessels in the region of interest (identified as the left hindlimb by in vivo imaging) were examined in further detail at an isotropic resolution of about 15 µm which revealed a vessel suspected of blockage. Subsequently, the limb of interest was isolated en bloc including the vessel of interest, for further characterisation of the site of suspected occlusion at a resolution of 3 µm by HREM and HP. HREM suggested a narrowing of the vessel lumen and evoked suspicion of intimal hyperplasia in the affected blood vessel segment within the VOI (
Figure 4). Only the inclusion of HREM in this pipeline allowed the detection of this narrowing of the vessel lumen and vascular intimal hyperplasia within the VOIs identified by CT and MRI [
16].
This entry is adapted from the peer-reviewed paper 10.3390/biomedicines9121918