CTCs as a potential biomarker of response to systemic therapies but also the technical challenges for their implementation in clinical practice, including the role of free circulating DNA and new approaches based on the isolation of CTCs from body fluids. New approaches focusing on isolation CTCs in other body fluids such as cerebrospinal or ascitic fluid are necessary to increase the opportunities of circulating tumor cells in the practice clinic as well as to study the promising role of CTC clusters and their prognostic value in metastatic breast cancer.
- CTCs
- breast cancer
- CTC clusters
- liquid biopsy
- body fluids
1. Introduction
The concept of tumor heterogeneity defines the existence at the same time of cellular subpopulations that differ from each other in their genetic and phenotypic characteristics, etc. This heterogeneity is also present between the primary tumor and its metastases. Among the causes that lead to this diversity are genetic and epigenetic factors and mechanisms such as adaptive responses, among others [1]. Techniques such as cytogenetic analysis, chromosomal analysis and microarray-based comparative genomic hybridization (CGH) have been used to demonstrate intra-tumor genetic heterogeneity in breast cancer [1]. Among the characteristics which conditionate the different subtypes, the expression of specific biomarkers, such as estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2—HER2, as well as different molecular profiles and tumor morphology are the most relevant [2]. Similarly, data suggest the existence of heterogeneity not only spatially (different subpopulations in different regions of the tumor) but also temporally (differences between the primary tumor and its recurrence) [1]. Thus, it should be taken into consideration that the sample obtained in a biopsy does not represent the totality of the composition of the tumor. The tumor is composed of different tumor cells that differ in their properties and drug resistance [2].
For this key reason, circulating tumor cells (CTCs) in peripheral blood provide the most objective way to assess intra tumor heterogeneity, facilitating the determination of single cell genotypes, cancer cell origin, differences among the CTCs and the solid tumor as well as those mutations which could address to the potential drug resistances (Figure 1). Thus, the CTCs detection together with optimized analyses methods are very helpful in the clinic for determining intra tumor heterogeneity [2].
TEPs are now considered to be local and systemic responders to the presence of cancer. They exert their effect both on tumor cells present in the tumor and on circulating tumor cells. Their mechanism of action is divided into several phases, first platelets generate an appropriate environment for neovascularization by providing the tumor with different proangiogenic factors such as VEGF, PDGF and FGF. Subsequently, platelets can induce epithelial-mesenchymal change in tumor cells by direct physical interaction and release of TGF molecules and have also been shown to reduce apoptosis and programmed cell death of local tumor cells. Once in the bloodstream, platelets are likely to protect circulating tumor cells from the immune response, favoring the metastatic process. Thus platelets are a fundamental component of the tumor microenvironment as they participate in tumor initiation, progression and response to therapy [4].
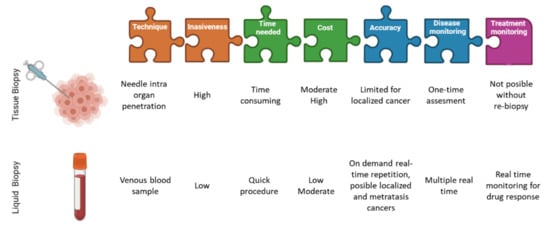
In the past few years, several clinical studies in breast and prostate cancer patients have highlighted the prognostic value of the detection and characterization of CTCs, suggesting they could even be categorized as follow-up markers and guide personalized treatment decisions. The idea of replacing tumor tissue biopsies to obtain diagnostically and therapeutically relevant information makes CTCs an essential contribution to non-invasive “real-time liquid biopsies” [5]. The next step in translating CTCs as liquid biopsy into clinical practice is to demonstrate the utility of these biomarkers. Observational clinical trials have already shown that CTCs and ctDNA are clinically relevant for different types of cancer. However, interventional clinical trials are needed to demonstrate their utility and to include liquid biopsies in clinical guidelines. Many clinical trials already include these determinations as part of specimen testing. However, the extent to which liquid biopsy could eventually replace tissue biopsies remains to be clarified. We know that for diagnosing primary tumors or staging metastatic lesions in tissues that are difficult to sample, liquid biopsy could be a reliable alternative [6]. Focusing on breast cancer, the biopsy of metastatic tissue is sometimes not clinically possible due to its location (lung) complicating the correctly determination of the molecular profile of a tumor by the limitation of taking biopsies from different tumor sites [6].
2. New Approaches
2.1. Optimize the Standardization of Protocols
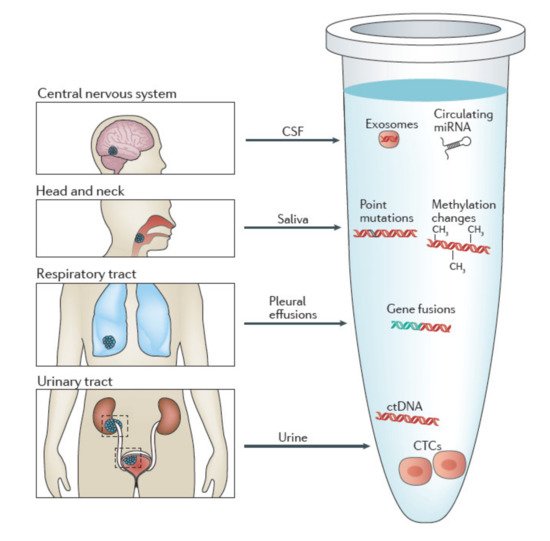
2.2. Isolation of CTCs in Other Body Fluids
-
Urine
-
Saliva
-
CSF
-
Other bodily fluids
2.3. Promising Role of CTC Clusters
This entry is adapted from the peer-reviewed paper 10.3390/cancers13184619
References
- Martelotto, L.G.; Ng, C.K.; Piscuoglio, S.; Weigelt, B.; Reis-Filho, J.S. Breast Cancer Intra-Tumor Heterogeneity. Breast Cancer Res. 2014, 16, 210.
- Januškevičienė, I.; Petrikaitė, V. Heterogeneity of Breast Cancer: The Importance of Interaction between Different Tumor Cell Populations. Life Sci. 2019, 239, 117009.
- Eslami-S., Z.; Cortés-Hernández, L.E.; Alix-Panabières, C. The Metastatic Cascade as the Basis for Liquid Biopsy Development. Front. Oncol. 2020, 10, 1055.
- Best, M.G.; Wesseling, P.; Wurdinger, T. Tumor-Educated Platelets as a Noninvasive Biomarker Source for Cancer Detection and Progression Monitoring. Cancer Res. 2018, 78, 3407–3412.
- Koch, C.; Kuske, A.; Joosse, S.A.; Yigit, G.; Sflomos, G.; Thaler, S.; Smit, D.J.; Werner, S.; Borgmann, K.; Gärtner, S.; et al. Characterization of Circulating Breast Cancer Cells with Tumorigenic and Metastatic Capacity. EMBO Mol. Med. 2020, 12, e11908.
- Alix-Panabières, C. The Future of Liquid Biopsy. Nature 2020, 579, S9.
- Alba-Bernal, A.; Lavado-Valenzuela, R.; Domínguez-Recio, M.E.; Jiménez-Rodriguez, B.; Queipo-Ortuño, M.I.; Alba, E.; Comino-Méndez, I. Challenges and Achievements of Liquid Biopsy Technologies Employed in Early Breast Cancer. EBioMedicine 2020, 62, 103100.
- Kowalik, A.; Kowalewska, M.; Góźdź, S. Current Approaches for Avoiding the Limitations of Circulating Tumor Cells Detection Methods—Implications for Diagnosis and Treatment of Patients with Solid Tumors. Transl. Res. 2017, 185, 58–84.e15.
- Arechederra, M.; Ávila, M.A.; Berasain, C. Liquid Biopsy for Cancer Management: A Revolutionary but Still Limited New Tool for Precision Medicine. Adv. Lab. Med. Av. Med. Lab. 2020, 1, 20200009.
- Bankó, P.; Lee, S.Y.; Nagygyörgy, V.; Zrínyi, M.; Chae, C.H.; Cho, D.H.; Telekes, A. Technologies for Circulating Tumor Cell Separation from Whole Blood. J. Hematol. Oncol. 2019, 12, 48.
- Bidard, F.-C.; Proudhon, C.; Pierga, J.-Y. Circulating Tumor Cells in Breast Cancer. Mol. Oncol. 2016, 10, 418–430.
- Neumann, M.H.D.; Bender, S.; Krahn, T.; Schlange, T. CtDNA and CTCs in Liquid Biopsy—Current Status and Where We Need to Progress. Comput. Struct. Biotechnol. J. 2018, 16, 190–195.
- Riethdorf, S.; Müller, V.; Loibl, S.; Nekljudova, V.; Weber, K.; Huober, J.; Fehm, T.; Schrader, I.; Hilfrich, J.; Holms, F.; et al. Prognostic Impact of Circulating Tumor Cells for Breast Cancer Patients Treated in the Neoadjuvant “Geparquattro” Trial. Clin. Cancer Res. 2017, 23, 5384–5393.
- Trapp, E.; Janni, W.; Schindlbeck, C.; Jückstock, J.; Andergassen, U.; de Gregorio, A.; Alunni-Fabbroni, M.; Tzschaschel, M.; Polasik, A.; Koch, J.G.; et al. Presence of Circulating Tumor Cells in High-Risk Early Breast Cancer During Follow-Up and Prognosis. J. Natl. Cancer Inst. 2019, 111, 380–387.
- Goodman, C.R.; Seagle, B.-L.L.; Friedl, T.W.P.; Rack, B.; Lato, K.; Fink, V.; Cristofanilli, M.; Donnelly, E.D.; Janni, W.; Shahabi, S.; et al. Association of Circulating Tumor Cell Status with Benefit of Radiotherapy and Survival in Early-Stage Breast Cancer. JAMA Oncol. 2018, 4, e180163.
- Kwan, T.T.; Bardia, A.; Spring, L.M.; Giobbie-Hurder, A.; Kalinich, M.; Dubash, T.; Sundaresan, T.; Hong, X.; LiCausi, J.A.; Ho, U.; et al. A Digital RNA Signature of Circulating Tumor Cells Predicting Early Therapeutic Response in Localized and Metastatic Breast Cancer. Cancer Discov. 2018, 8, 1286–1299.
- Bünger, S.; Zimmermann, M.; Habermann, J.K. Diversity of Assessing Circulating Tumor Cells (CTCs) Emphasizes Need for Standardization: A CTC Guide to Design and Report Trials. Cancer Metastasis Rev. 2015, 34, 527–545.
- De Rubis, G.; Rajeev Krishnan, S.; Bebawy, M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol. Sci. 2019, 40, 172–186.
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating Liquid Biopsies into the Management of Cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548.
- Oshi, M.; Murthy, V.; Takahashi, H.; Huyser, M.; Okano, M.; Tokumaru, Y.; Rashid, O.M.; Matsuyama, R.; Endo, I.; Takabe, K. Urine as a Source of Liquid Biopsy for Cancer. Cancers 2021, 13, 2652.
- Husain, H.; Nykin, D.; Bui, N.; Quan, D.; Gomez, G.; Woodward, B.; Venkatapathy, S.; Duttagupta, R.; Fung, E.; Lippman, S.M.; et al. Cell-Free DNA from Ascites and Pleural Effusions: Molecular Insights into Genomic Aberrations and Disease Biology. Mol. Cancer Ther. 2017, 16, 948–955.
- Su, Y.-H.; Wang, M.; Brenner, D.E.; Norton, P.A.; Block, T.M. Detection of Mutated K- Ras DNA in Urine, Plasma, and Serum of Patients with Colorectal Carcinoma or Adenomatous Polyps. Ann. N. Y. Acad. Sci. 2008, 1137, 197–206.
- Iwasaki, H.; Shimura, T.; Yamada, T.; Okuda, Y.; Natsume, M.; Kitagawa, M.; Horike, S.; Kataoka, H. A Novel Urinary MicroRNA Biomarker Panel for Detecting Gastric Cancer. J. Gastroenterol. 2019, 54, 1061–1069.
- Zhang, J.; Zhang, X.; Shen, S. Treatment and Relapse in Breast Cancer Show Significant Correlations to Noninvasive Testing Using Urinary and Plasma DNA. Future Oncol. 2020, 16, 849–858.
- Dhondt, B.; Van Deun, J.; Vermaerke, S.; de Marco, A.; Lumen, N.; De Wever, O.; Hendrix, A. Urinary Extracellular Vesicle Biomarkers in Urological Cancers: From Discovery towards Clinical Implementation. Int. J. Biochem. Cell Biol. 2018, 99, 236–256.
- Lin, S.-Y.; Chang, C.-H.; Wu, H.-C.; Lin, C.-C.; Chang, K.-P.; Yang, C.-R.; Huang, C.-P.; Hsu, W.-H.; Chang, C.-T.; Chen, C.-J. Proteome Profiling of Urinary Exosomes Identifies Alpha 1-Antitrypsin and H2B1K as Diagnostic and Prognostic Biomarkers for Urothelial Carcinoma. Sci. Rep. 2016, 6, 34446.
- Streckfus, C.; Bigler, L. The Use of Soluble, Salivary c- ErbB-2 for the Detection and Post-Operative Follow-up of Breast Cancer in Women: The Results of a Five-Year Translational Research Study. Adv. Dent. Res. 2005, 18, 17–24.
- Lau, C.S.; Wong, D.T.W. Breast Cancer Exosome-like Microvesicles and Salivary Gland Cells Interplay Alters Salivary Gland Cell-Derived Exosome-like Microvesicles In Vitro. PLoS ONE 2012, 7, e33037.
- Zhang, L.; Xiao, H.; Karlan, S.; Zhou, H.; Gross, J.; Elashoff, D.; Akin, D.; Yan, X.; Chia, D.; Karlan, B.; et al. Discovery and Preclinical Validation of Salivary Transcriptomic and Proteomic Biomarkers for the Non-Invasive Detection of Breast Cancer. PLoS ONE 2010, 5, e15573.
- Seoane, J.; De Mattos-Arruda, L.; Le Rhun, E.; Bardelli, A.; Weller, M. Cerebrospinal Fluid Cell-Free Tumour DNA as a Liquid Biopsy for Primary Brain Tumours and Central Nervous System Metastases. Ann. Oncol. 2019, 30, 211–218.
- Wang, C.; Mu, Z.; Chervoneva, I.; Austin, L.; Ye, Z.; Rossi, G.; Palazzo, J.P.; Sun, C.; Abu-Khalaf, M.; Myers, R.E.; et al. Longitudinally Collected CTCs and CTC-Clusters and Clinical Outcomes of Metastatic Breast Cancer. Breast Cancer Res. Treat. 2017, 161, 83–94.
- Piñeiro, R.; Martínez-Pena, I.; López-López, R. Relevance of CTC Clusters in Breast Cancer Metastasis. Adv. Exp. Med. Biol. 2020, 1220, 93–115.
- Yang, C.; Xia, B.-R.; Jin, W.-L.; Lou, G. Circulating Tumor Cells in Precision Oncology: Clinical Applications in Liquid Biopsy and 3D Organoid Model. Cancer Cell Int. 2019, 19, 341.
- Fabisiewicz, A.; Szostakowska-Rodzos, M.; Zaczek, A.J.; Grzybowska, E.A. Circulating Tumor Cells in Early and Advanced Breast Cancer; Biology and Prognostic Value. Int. J. Mol. Sci. 2020, 21, 1671.
- Menyailo, M.E.; Tretyakova, M.S.; Denisov, E.V. Heterogeneity of Circulating Tumor Cells in Breast Cancer: Identifying Metastatic Seeds. Int. J. Mol. Sci. 2020, 21, 1696.
- Padmanaban, V.; Krol, I.; Suhail, Y.; Szczerba, B.M.; Aceto, N.; Bader, J.S.; Ewald, A.J. E-Cadherin Is Required for Metastasis in Multiple Models of Breast Cancer. Nature 2019, 573, 439–444.
- Krol, I.; Schwab, F.D.; Carbone, R.; Ritter, M.; Picocci, S.; De Marni, M.L.; Stepien, G.; Franchi, G.M.; Zanardi, A.; Rissoglio, M.D.; et al. Detection of Clustered Circulating Tumour Cells in Early Breast Cancer. Br. J. Cancer 2021, 125, 23–27.
- Au, S.H.; Edd, J.; Stoddard, A.E.; Wong, K.H.K.; Fachin, F.; Maheswaran, S.; Haber, D.A.; Stott, S.L.; Kapur, R.; Toner, M. Microfluidic Isolation of Circulating Tumor Cell Clusters by Size and Asymmetry. Sci. Rep. 2017, 7, 2433.
- Gkountela, S.; Castro-Giner, F.; Szczerba, B.M.; Vetter, M.; Landin, J.; Scherrer, R.; Krol, I.; Scheidmann, M.C.; Beisel, C.; Stirnimann, C.U.; et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell 2019, 176, 98–112.e14.
