Ulcerative colitis (UC) is a major type of inflammatory bowel disease (IBD), characterized by chronic inflammation of the colon and rectum. Inflammation confined to the mucosa is distributed continuously from the rectum to the proximal colon in UC and causes bloody stools, diarrhea, and abdominal pain.
- ulcerative colitis
- inflammatory bowel disease
- microRNA
1. Introduction
Studies have demonstrated that the etiology of UC is based on complicated interactions among intestinal microbiota, dietary components, and host immune systems in genetically susceptible patients. However, the precise mechanisms underlying this pathology remain unclear [1]. Although there is currently no cure for UC, several new immunomodulatory drugs have been developed in recent decades. Furthermore, recent progress in genome-wide association studies (GWAS) has led to the identification of disease-associated loci, highlighting the mechanistic pathways involved in the development of UC, such as epithelial barrier, innate mucosal defense, and adaptive immune cell regulation [2]. However, the association risk with individual susceptibility loci is small and GWAS signals are often located in non-coding regions of the genome, suggesting that protein-coding genes alone cannot explain the disease mechanism. As a result, subsequent research has investigated the role of non-coding RNA, including microRNA in the pathophysiology of UC.
MicroRNAs (miRNAs) are a group of small (~22 nucleotides) non-coding RNAs that confer post-transcriptional regulation of target gene expression. Each miRNA can target hundreds of gene transcripts. Over 60% of human protein-coding genes harbor predicted miRNA target sites [3] that participate in the regulation of various biological processes, including cell proliferation, differentiation, apoptosis, and signal transduction [4]. As a result, miRNAs are implicated in a variety of diseases, including cancer, neurological diseases, cardiovascular diseases, and autoimmune diseases, such as IBD [5][6]. Dysregulated miRNA expression profiles have been reported in the saliva, peripheral blood, and intestinal mucosa of patients with UC compared to healthy individuals and patients with Crohn’s disease (CD), another type of IBD [7][8].
2. miRNA Profile in UC
Since first being reported in 2008 [9], an increasing number of studies have identified dysregulated miRNA expression profiles in the saliva, peripheral blood, and colonic mucosa of patients with UC [10]. These studies, especially those focusing on body fluids, were partially motivated by the potential usefulness of miRNAs as biomarkers for the diagnosis of disease or the monitoring of disease activity in UC. However, it should be noted that the miRNA profiles in the colon are different from those in body fluids [11]. In addition, heterogeneous comparators, including healthy individuals, patients with CD, and other types of intestinal disorders included in these studies make it difficult to interpret the significance of dysregulated miRNAs in the pathophysiology of UC.
3. miRNAs in the Pathophysiology of UC
Intestinal homeostasis is maintained on a single layer of epithelial cells, which separates potentially harmful luminal antigens, such as dietary components and microbiota, from mucosal immune cells. The mucus produced by goblet cells overlying the epithelium provides additional protection against the invasion of these antigens. The breakdown of these epithelial barrier systems and the subsequent activation of mucosal immune cells are critical steps in the disruption of intestinal homeostasis and disease development in UC.
The importance of miRNAs in epithelial barrier function has been previously reported in intestinal epithelial cell (IEC)-specific Dicer1 knock-out (KO) mice, which lack all miRNAs in IECs [12]. Increased epithelial permeability and the development of spontaneous intestinal inflammation in these mice suggest a critical role for miRNAs in IEC biology and maintenance of epithelial barrier integrity. Similarly, selective Dicer1 or Drosha deletion in immune cells implicates miRNAs in regulating immune cell functions. Bone marrow-derived macrophages from Dicer1 KO mice produce decreased levels of pro-inflammatory cytokines compared to control mice upon stimulation with Toll-like receptors (TLRs) [13]. Dicer1-deficient T cells preferentially produce interferon gamma (IFN-γ) [14], while Drosha deficiency impairs the suppressive ability of regulatory T cells (Tregs) and induces spontaneous inflammatory disease in mice [15]. These findings collectively indicate the importance of miRNAs in maintaining intestinal homeostasis and suggest their potential roles in the pathophysiology of UC.
miRNAs are also involved in macrophage polarization, a process by which macrophages adopt distinct functional phenotypes in response to signals from the microenvironment ( Figure 1 ) [16]. Classically activated (M1) macrophages have a pro-inflammatory phenotype, whereas alternatively activated (M2) macrophages have an anti-inflammatory phenotype and are involved in tissue repair [17]. MiR-125b and miR-155 drive M1 polarization by directly targeting interferon regulatory factor 4 (Irf4) and IL13RA1, respectively [18][19]. MiR-146a drives M2 polarization, at least in part, by modulating the expression of Notch1 and inhibin β A subunit of activin A (INHBA) [20][21]. MiR-223 is required for peroxisome proliferator-activated receptor γ (PPARγ)-dependent M2 macrophage activation by targeting nuclear factor of activated T cells 5 (Nfat5) and Ras p21 protein activator 1 (Rasa1) in mice [22]. Collectively, miRNAs play a crucial role in regulating innate immune responses by targeting PRR signaling molecules at multiple levels of cascades and directly modulating the functions of innate immune cells.
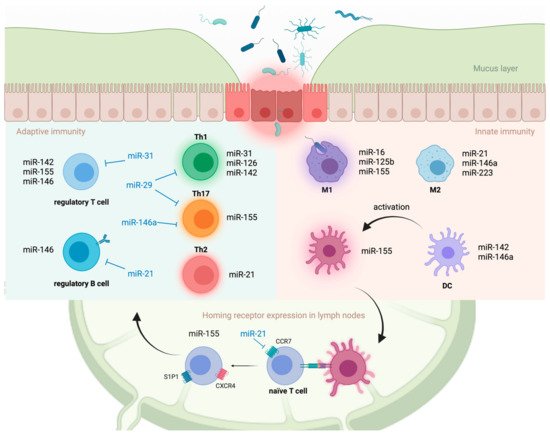
Antigen presentation and cytokine production by activated innate immune cells induce the maturation and development of T cells within inductive sites, such as gut-associated lymphoid tissue (GALT) and mesenteric lymph nodes. Aberrant T cell activation and preferential differentiation towards pathogenic phenotypes on these inductive sites are strongly implicated in the development of UC [23]. Although UC is thought to be a type 2 helper T (Th2) cell-driven disease, studies have demonstrated the involvement of other T cell lineages, including Th1, Th9, Th17, and Tregs, in disease progression [23].
4. Potential of miRNAs as Therapeutic Targets in UC
There is a growing interest in the manipulation of miRNAs with antagonists or mimics for use as therapeutic targets in UC, similar to other fields of disease [24][25][26]. However, despite promising results from rodent models of IBD, clinical application of miRNA-based therapy has not yet been established in UC due to several limitations [27]. First, currently available data on miRNA expression profiles in patients with UC are heterogeneous in terms of clinical background. The potential impacts of a heterogeneous background, including disease activity, duration, and treatment contents, on miRNA expression should be noted when identifying a therapeutic target of miRNA. Second, previous reports have investigated colonic miRNA expression profiles in diseases endoscopically using biopsy specimens. Therefore, the cell-type specific expression of miRNAs is largely unknown. Investigating miRNA expression in a single cell-type specific manner is necessary to understand the impact of dysregulated miRNAs on the etiology of UC and to identify appropriate targets for therapeutic manipulation. Third, recent advances in technologies of drug delivery, such as lipid nanoparticles and dendrimer complexes with a targeting moiety attached, have enabled miRNA-based treatment in clinical practice [28]. These include miR-16 mimic (MesomiR-1, EnGeneIC Ltd., Sydney, NSW, Australia) encapsulated in lipid vesicles for mesothelioma [29], cholesterol-conjugated miR-29 mimic ( Remlarsen/MRG-201, miRagen Therapeutics Inc., Boulder, CO, USA) for scleroderma [30], and a locked nucleic acid-modified oligonucleotide inhibitor of miR-155 (Cobomarsen/MRG-106, miRagen Therapeutics Inc., Boulder, CO, USA) for cutaneous T cell lymphoma [31]. Despite these advances, tissue- or cell type-specific treatments remain technically challenging. Since individual miRNAs can target hundreds of transcripts, establishing a tissue/cell type-specific targeting technology is key to avoid potential toxicities and off-target effects. Successfully resolving these problems will carve a concrete path toward the development of novel miRNA-based therapeutics in UC, which would enhance intestinal epithelial barrier and/or modulate mucosal immune response. Antagonizing miR-21 and miR-155 might be of particular interest for clinical application considering their multifaceted roles in the pathophysiology of UC.
This entry is adapted from the peer-reviewed paper 10.3390/immuno1040039
References
- Sartor, R.B. Mechanisms of disease: Pathogenesis of Crohn’s disease and ulcerative colitis. Nat. Clin. Pract. Gastroenterol. Hepatol. 2006, 3, 390–407.
- Khor, B.; Gardet, A.; Xavier, R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011, 474, 307–317.
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105.
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51.
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874.
- Kalla, R.; Ventham, N.T.; Kennedy, N.A.; Quintana, J.F.; Nimmo, E.R.; Buck, A.H.; Satsangi, J. MicroRNAs: New players in IBD. Gut 2015, 64, 504–517.
- Zhou, J.; Liu, J.; Gao, Y.; Shen, L.; Li, S.; Chen, S. miRNA-Based Potential Biomarkers and New Molecular Insights in Ulcerative Colitis. Front. Pharmacol. 2021, 12, 707776.
- Schaefer, J.S.; Attumi, T.; Opekun, A.R.; Abraham, B.; Hou, J.; Shelby, H.; Graham, D.Y.; Streckfus, C.; Klein, J.R. MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC Immunol. 2015, 16, 5.
- Wu, F.; Zikusoka, M.; Trindade, A.; Dassopoulos, T.; Harris, M.L.; Bayless, T.M.; Brant, S.R.; Chakravarti, S.; Kwon, J.H. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology 2008, 135, 1624–1635.e24.
- Ghafouri-Fard, S.; Eghtedarian, R.; Taheri, M. The crucial role of non-coding RNAs in the pathophysiology of inflammatory bowel disease. Biomed. Pharmacother. 2020, 129, 110507.
- Iborra, M.; Bernuzzi, F.; Correale, C.; Vetrano, S.; Fiorino, G.; Beltran, B.; Marabita, F.; Locati, M.; Spinelli, A.; Nos, P.; et al. Identification of serum and tissue micro-RNA expression profiles in different stages of inflammatory bowel disease. Clin. Exp. Immunol. 2013, 173, 250–258.
- McKenna, L.B.; Schug, J.; Vourekas, A.; McKenna, J.B.; Bramswig, N.C.; Friedman, J.R.; Kaestner, K.H. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology 2010, 139, 1654–1664.e1.
- Gantier, M.P.; Stunden, H.J.; McCoy, C.E.; Behlke, M.A.; Wang, D.; Kaparakis-Liaskos, M.; Sarvestani, S.T.; Yang, Y.H.; Xu, D.; Corr, S.C.; et al. A miR-19 regulon that controls NF-kappaB signaling. Nucleic Acids Res. 2012, 40, 8048–8058.
- Muljo, S.A.; Ansel, K.M.; Kanellopoulou, C.; Livingston, D.M.; Rao, A.; Rajewsky, K. Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 2005, 202, 261–269.
- Chong, M.M.; Rasmussen, J.P.; Rudensky, A.Y.; Littman, D.R. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J. Exp. Med. 2008, 205, 2005–2017.
- Curtale, G.; Rubino, M.; Locati, M. MicroRNAs as Molecular Switches in Macrophage Activation. Front. Immunol. 2019, 10, 799.
- Italiani, P.; Boraschi, D. From Monocytes to M1/M2 Macrophages: Phenotypical vs. Functional Differentiation. Front. Immunol. 2014, 5, 514.
- Chaudhuri, A.A.; So, A.Y.; Sinha, N.; Gibson, W.S.; Taganov, K.D.; O’Connell, R.M.; Baltimore, D. MicroRNA-125b potentiates macrophage activation. J. Immunol. 2011, 187, 5062–5068.
- Martinez-Nunez, R.T.; Louafi, F.; Sanchez-Elsner, T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor alpha1 (IL13Ralpha1). J. Biol. Chem. 2011, 286, 1786–1794.
- Huang, C.; Liu, X.J.; QunZhou; Xie, J.; Ma, T.T.; Meng, X.M.; Li, J. MiR-146a modulates macrophage polarization by inhibiting Notch1 pathway in RAW264.7 macrophages. Int. Immunopharmacol. 2016, 32, 46–54.
- Li, D.; Duan, M.; Feng, Y.; Geng, L.; Li, X.; Zhang, W. Corrigendum to “MiR-146a modulates macrophage polarization in systemic juvenile idiopathic arthritis by targeting INHBA” . Mol. Immunol. 2017, 87, 329–330.
- Ying, W.; Tseng, A.; Chang, R.C.; Morin, A.; Brehm, T.; Triff, K.; Nair, V.; Zhuang, G.; Song, H.; Kanameni, S.; et al. MicroRNA-223 is a crucial mediator of PPARgamma-regulated alternative macrophage activation. J. Clin. Investig. 2015, 125, 4149–4159.
- Choy, M.C.; Visvanathan, K.; De Cruz, P. An Overview of the Innate and Adaptive Immune System in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2017, 23, 2–13.
- Titze-de-Almeida, S.S.; Soto-Sanchez, C.; Fernandez, E.; Koprich, J.B.; Brotchie, J.M.; Titze-de-Almeida, R. The Promise and Challenges of Developing miRNA-Based Therapeutics for Parkinson’s Disease. Cells 2020, 9, 841.
- Naidu, S.; Magee, P.; Garofalo, M. MiRNA-based therapeutic intervention of cancer. J. Hematol. Oncol. 2015, 8, 68.
- Pang, J.K.S.; Phua, Q.H.; Soh, B.S. Applications of miRNAs in cardiac development, disease progression and regeneration. Stem Cell Res. Ther. 2019, 10, 336.
- Suri, K.; Bubier, J.A.; Wiles, M.V.; Shultz, L.D.; Amiji, M.M.; Hosur, V. Role of MicroRNA in Inflammatory Bowel Disease: Clinical Evidence and the Development of Preclinical Animal Models. Cells 2021, 10, 2204.
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222.
- Van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017, 18, 1386–1396.
- Gallant-Behm, C.L.; Piper, J.; Lynch, J.M.; Seto, A.G.; Hong, S.J.; Mustoe, T.A.; Maari, C.; Pestano, L.A.; Dalby, C.M.; Jackson, A.L.; et al. A MicroRNA-29 Mimic (Remlarsen) Represses Extracellular Matrix Expression and Fibroplasia in the Skin. J. Investig. Dermatol. 2019, 139, 1073–1081.
- Seto, A.G.; Beatty, X.; Lynch, J.M.; Hermreck, M.; Tetzlaff, M.; Duvic, M.; Jackson, A.L. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br. J. Haematol. 2018, 183, 428–444.
