Sphingolipids are ubiquitous components of cellular membranes that exert various functions depending on their structural maturation and subcellular localization. Structurally simple sphingolipid precursors, such as ceramides, act as intracellular signaling molecules in many processes, including apoptosis, whereas mature and complex forms of sphingolipids are important structural components of the plasma membrane. Supplying complex sphingolipids to the plasma membrane while simultaneously preventing the accumulation of pro-apoptotic metabolites is essential for cell survival and depends on mechanisms that tightly control sphingolipid synthesis, breakdown, transport, and storage. Sphingolipid homeostasis describes the state of the cell in which the intracellular concentration and distribution of sphingolipids supports survival.
- sphingolipids
- membrane contact sites
- metabolism
1. Introduction
2. Sphingolipid Synthesis, Catabolism, and Trafficking in Yeast
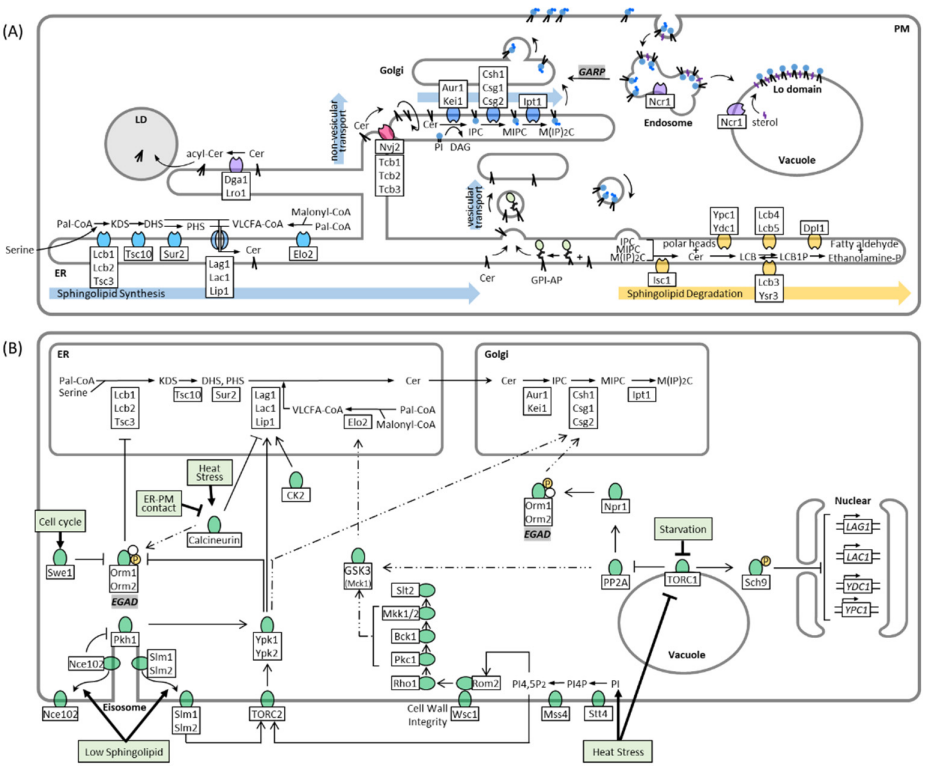
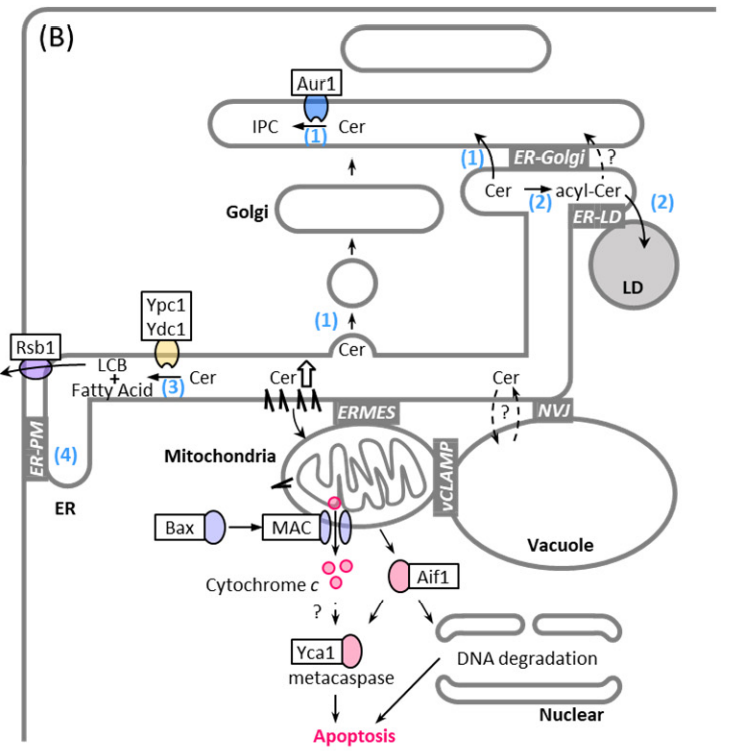
3. Regulation of Sphingolipid Metabolism
This entry is adapted from the peer-reviewed paper 10.3390/membranes11120971
References
- Ziółkowska, N.E.; Christiano, R.; Walther, T.C. Organized living: Formation mechanisms and functions of plasma membrane domains in yeast. Trends Cell Biol. 2012, 22, 151–158.
- Hannun, Y.A.; Obeid, L.M. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150.
- Pettus, B.J.; Chalfant, C.E.; Hannun, Y.A. Ceramide in apoptosis: An overview and current perspectives. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2002, 1585, 114–125.
- Mandala, S.M.; Thornton, R.; Tu, Z.; Kurtz, M.B.; Nickels, J.; Broach, J.; Menzeleev, R.; Spiegel, S. Sphingoid base 1-phosphate phosphatase: A key regulator of sphingolipid metabolism and stress response. Proc Natl. Acad. Sci. USA 1998, 95, 150–155.
- Morad, S.A.; Cabot, M.C. Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer 2013, 13, 51–65.
- Shimobayashi, M.; Oppliger, W.; Moes, S.; Jenö, P.; Hall, M.N. TORC1-regulated protein kinase Npr1 phosphorylates Orm to stimulate complex sphingolipid synthesis. Mol. Biol. Cell 2013, 24, 870–881.
- Berchtold, D.; Piccolis, M.; Chiaruttini, N.; Riezman, I.; Riezman, H.; Roux, A.; Walther, T.; Loewith, R. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat. Cell Biol. 2012, 14, 542–547.
- Gururaj, C.; Federman, R.; Chang, A. Orm proteins integrate multiple signals to maintain sphingolipid homeostasis. J. Biol. Chem. 2013, 288, 20453–20463.
- Olson, D.K.; Fröhlich, F.; Farese, R.V., Jr.; Walther, T.C. Taming the sphinx: Mechanisms of cellular sphingolipid homeostasis. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2016, 1861, 784–792.
- Senkal, C.E.; Salama, M.F.; Snider, A.J.; Allopenna, J.J.; Rana, N.A.; Koller, A.; Hannun, Y.A.; Obeid, L.M. Ceramide is metabolized to acylceramide and stored in lipid droplets. Cell Metab. 2017, 25, 686–697.
- Voynova, N.S.; Vionnet, C.; Ejsing, C.S.; Conzelmann, A. A novel pathway of ceramide metabolism in Saccharomyces cerevisiae. Biochem. J. 2012, 447, 103–114.
- Ikeda, A.; Schlarmann, P.; Kurokawa, K.; Nakano, A.; Riezman, H.; Funato, K. Tricalbins are required for non-vesicular ceramide transport at ER-Golgi contacts and modulate lipid droplet biogenesis. Iscience 2020, 23, 101603.
- Mandon, E.C.; Ehses, I.; Rother, J.; van Echten, G.; Sandhoff, K. Subcellular localization and membrane topology of serine palmitoyltransferase, 3-dehydrosphinganine reductase, and sphinganine N-acyltransferase in mouse liver. J. Biol. Chem. 1992, 267, 11144–11148.
- Han, G.; Gable, K.; Yan, L.; Natarajan, M.; Krishnamurthy, J.; Gupta, S.D.; Borovitskaya, A.; Harmon, J.M.; Dunn, T.M. The topology of the Lcb1p subunit of yeast serine palmitoyltransferase. J. Biol. Chem. 2004, 279, 53707–53716.
- Gable, K.; Han, G.; Monaghan, E.; Bacikova, D.; Natarajan, M.; Williams, R.; Dunn, T.M. Mutations in the yeast LCB1 and LCB2Genes, including those corresponding to the hereditary sensory neuropathy type I mutations, dominantly inactivate serine palmitoyltransferase. J. Biol. Chem. 2002, 277, 10194–10200.
- Kihara, A.; Igarashi, Y. FVT-1 is a mammalian 3-ketodihydrosphingosine reductase with an active site that faces the cytosolic side of the endoplasmic reticulum membrane. J. Biol. Chem. 2004, 279, 49243–49250.
- Kageyama-Yahara, N.; Riezman, H. Transmembrane topology of ceramide synthase in yeast. Biochem. J. 2006, 398, 585–593.
- Megyeri, M.; Prasad, R.; Volpert, G.; Sliwa-Gonzalez, A.; Haribowo, A.G.; Aguilera-Romero, A.; Riezman, H.; Barral, Y.; Futerman, A.; Schuldiner, M. Yeast ceramide synthases, Lag1 and Lac1, have distinct substrate specificity. J. Cell Sci. 2019, 132, jcs228411.
- Vallée, B.; Riezman, H. Lip1p: A novel subunit of acyl-CoA ceramide synthase. EMBO J. 2005, 24, 730–741.
- Funato, K.; Lombardi, R.; Vallée, B.; Riezman, H. Lcb4p is a key regulator of ceramide synthesis from exogenous long chain sphingoid base in Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 7325–7334.
- Kihara, A.; Sano, T.; Iwaki, S.; Igarashi, Y. Transmembrane topology of sphingoid long-chain base-1-phosphate phosphatase, Lcb3p. Genes Cells 2003, 8, 525–535.
- Funato, K.; Vallée, B.; Riezman, H. Biosynthesis and trafficking of sphingolipids in the yeast Saccharomyces cerevisiae. Biochemistry 2002, 41, 15105–15114.
- Kondo, N.; Ohno, Y.; Yamagata, M.; Obara, T.; Seki, N.; Kitamura, T.; Naganuma, T.; Kihara, A. Identification of the phytosphingosine metabolic pathway leading to odd-numbered fatty acids. Nat. Commun. 2014, 5, 5338.
- Mayor, S.; Riezman, H. Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 2004, 5, 110–120.
- Bosson, R.; Guillas, I.; Vionnet, C.; Roubaty, C.; Conzelmann, A. Incorporation of ceramides into Saccharomyces cerevisiae glycosylphosphatidylinositol-anchored proteins can be monitored in vitro. Eukaryot. Cell 2009, 8, 306–314.
- Swain, E.; Stukey, J.; McDonough, V.; Germann, M.; Liu, Y.; Sturley, S.L.; Nickels, J.T. Yeast cells lacking the ARV1 gene harbor defects in sphingolipid metabolism: Complementation by human ARV1. J. Biol. Chem. 2002, 277, 36152–36160.
- Kajiwara, K.; Watanabe, R.; Pichler, H.; Ihara, K.; Murakami, S.; Riezman, H.; Funato, K. Yeast ARV1 is required for efficient delivery of an early GPI intermediate to the first mannosyltransferase during GPI assembly and controls lipid flow from the endoplasmic reticulum. Mol. Biol. Cell 2008, 19, 2069–2082.
- Sutterlin, C.; Doering, T.L.; Schimmoller, F.; Schroder, S.; Riezman, H. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J. Cell Sci. 1997, 110, 2703–2714.
- Bagnat, M.; Keränen, S.; Shevchenko, A.; Shevchenko, A.; Simons, K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Natl. Acad. Sci. USA 2000, 97, 3254–3259.
- Rodriguez-Gallardo, S.; Kurokawa, K.; Sabido-Bozo, S.; Cortes-Gomez, A.; Ikeda, A.; Zoni, V.; Aguilera-Romero, A.; Perez-Linero, A.M.; Lopez, S.; Waga, M.; et al. Ceramide chain length–dependent protein sorting into selective endoplasmic reticulum exit sites. Sci. Adv. 2020, 6, eaba8237.
- Funato, K.; Riezman, H. Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J. Cell Biol. 2001, 155, 949–960.
- Liu, L.K.; Choudhary, V.; Toulmay, A.; Prinz, W.A. An inducible ER–Golgi tether facilitates ceramide transport to alleviate lipotoxicity. J. Cell Biol. 2017, 216, 131–147.
- Hanada, K.; Kumagai, K.; Yasuda, S.; Miura, Y.; Kawano, M.; Fukasawa, M.; Nishijima, M. Molecular machinery for non-vesicular trafficking of ceramide. Nature 2003, 426, 803–809.
- Kumagai, K.; Hanada, K. Structure, functions and regulation of CERT, a lipid-transfer protein for the delivery of ceramide at the ER-Golgi membrane contact sites. FEBS Lett. 2019, 593, 2366–2377.
- Levine, T.P.; Wiggins, C.A.; Munro, S. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell 2000, 11, 2267–2281.
- Sato, K.; Noda, Y.; Yoda, K. Kei1: A novel subunit of inositolphosphorylceramide synthase, essential for its enzyme activity and Golgi localization. Mol. Biol. Cell 2009, 20, 4444–4457.
- Lisman, Q.; Pomorski, T.; Vogelzangs, C.; Urli-Stam, D.; van Delwijnen, W.D.C.; Holthuis, J.C. Protein sorting in the late Golgi of Saccharomyces cerevisiae does not require mannosylated sphingolipids. J. Biol. Chem. 2004, 279, 1020–1029.
- Uemura, S.; Kihara, A.; Inokuchi, J.I.; Igarashi, Y. Csg1p and newly identified Csh1p function in mannosylinositol phosphorylceramide synthesis by interacting with Csg2p. J. Biol. Chem. 2003, 278, 45049–45055.
- Dickson, R.C.; Nagiec, E.E.; Wells, G.B.; Nagiec, M.M.; Lester, R.L. Synthesis of mannose-(inositol-P) 2-ceramide, the major sphingolipid in Saccharomyces cerevisiae, requires the IPT1 (YDR072c) gene. J. Biol. Chem. 1997, 272, 29620–29625.
- Schnabl, M.; Daum, G.; Pichler, H. Multiple lipid transport pathways to the plasma membrane in yeast. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2005, 1687, 130–140.
- Sawai, H.; Okamoto, Y.; Luberto, C.; Mao, C.; Bielawska, A.; Domae, N.; Hannun, Y.A. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 39793–39798.
- De Avalos, S.V.; Okamoto, Y.; Hannun, Y.A. Activation and localization of inositol phosphosphingolipid phospholipase C, Isc1p, to the mitochondria during growth of Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 11537–11545.
- Ohno, Y.; Kamiyama, N.; Nakamichi, S.; Kihara, A. PNPLA1 is a transacylase essential for the generation of the skin barrier lipid ω-O-acylceramide. Nat. Commun. 2017, 8, 14610.
- Klug, L.; Daum, G. Yeast lipid metabolism at a glance. FEMS Yeast Res. 2014, 14, 369–388.
- Breslow, D.K. Sphingolipid homeostasis in the endoplasmic reticulum and beyond. Cold Spring Harb. Perspect. Biol. 2013, 5, a013326.
- Platt, F.M. Sphingolipid lysosomal storage disorders. Nature 2014, 510, 68–75.
- Teixeira, V.; Medeiros, T.C.; Vilaça, R.; Ferreira, J.; Moradas-Ferreira, P.; Costa, V. Ceramide signaling targets the PP2A-like protein phosphatase Sit4p to impair vacuolar function, vesicular trafficking and autophagy in Isc1p deficient cells. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2016, 1861, 21–33.
- Hurst, L.R.; Fratti, R.A. Lipid rafts, sphingolipids, and ergosterol in yeast vacuole fusion and maturation. Front. Cell Dev. Biol. 2020, 8, 539.
- Barbosa, A.D.; Osório, H.; Sims, K.J.; Almeida, T.; Alves, M.; Bielawski, J.; Amorim, M.A.; Moradas-Ferreira, P.; Hannun, Y.A.; Costa, V. Role for Sit4p-dependent mitochondrial dysfunction in mediating the shortened chronological lifespan and oxidative stress sensitivity of Isc1p-deficient cells. Mol. Microbiol. 2011, 81, 515–527.
- Malathi, K.; Higaki, K.; Tinkelenberg, A.H.; Balderes, D.A.; Almanzar-Paramio, D.; Wilcox, L.J.; Erdeniz, N.; Redican, F.; Padamsee, M.; Liu, Y.; et al. Mutagenesis of the putative sterol-sensing domain of yeast Niemann Pick C–related protein reveals a primordial role in subcellular sphingolipid distribution. J. Cell Biol. 2004, 164, 547–556.
- Conibear, E.; Cleck, J.N.; Stevens, T.H. Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol. Biol. Cell 2003, 14, 1610–1623.
- Fröhlich, F.; Petit, C.; Kory, N.; Christiano, R.; Hannibal-Bach, H.K.; Graham, M.; Liu, X.; Ejsing, C.S.; Farese, R.V.; Walther, T.C. The GARP complex is required for cellular sphingolipid homeostasis. eLife 2015, 4, e08712.
- Tsuji, T.; Fujimoto, M.; Tatematsu, T.; Cheng, J.; Orii, M.; Takatori, S.; Fujimoto, T. Niemann-Pick type C proteins promote microautophagy by expanding raft-like membrane domains in the yeast vacuole. eLife 2017, 6, e25960.
- Kihara, A.; Igarashi, Y. Identification and characterization of a Saccharomyces cerevisiae gene, RSB1, involved in sphingoid long-chain base release. J. Biol. Chem. 2002, 277, 30048–30054.
- Heidler, S.A.; Radding, J.A. The AUR1 gene in Saccharomyces cerevisiae encodes dominant resistance to the antifungal agent aureobasidin A (LY295337). Antimicrob. Agents Chemother. 1995, 39, 2765–2769.
- Hashida-Okado, T.; Ogawa, A.; Endo, M.; Yasumoto, R.; Takesako, K.; Kato, I. AUR1, a novel gene conferring aureobasidin resistance on Saccharomyces cerevisiae: A study of defective morphologies in Aur1p-depleted cells. Mol. Gen. Genet. 1996, 251, 236–244.
- Schorling, S.; Vallée, B.; Barz, W.P.; Riezman, H.; Oesterhelt, D. Lag1p and Lac1p are essential for the Acyl-CoA–dependent ceramide synthase reaction in Saccharomyces cerevisae. Mol. Biol. Cell 2001, 12, 3417–3427.
- Kajiwara, K.; Muneoka, T.; Watanabe, Y.; Karashima, T.; Kitagaki, H.; Funato, K. Perturbation of sphingolipid metabolism induces endoplasmic reticulum stress-mediated mitochondrial apoptosis in budding yeast. Mol. Microbiol. 2012, 86, 1246–1261.
- Eisenberg, T.; Büttner, S. Lipids and cell death in yeast. FEMS Yeast Res. 2014, 14, 179–197.
- Rego, A.; Duarte, A.M.; Azevedo, F.; Sousa, M.J.; Côrte-Real, M.; Chaves, S.R. Cell wall dynamics modulate acetic acid-induced apoptotic cell death of Saccharomyces cerevisiae. Microb. Cell 2014, 1, 303.
- Voynova, N.S.; Roubaty, C.; Vazquez, H.M.; Mallela, S.K.; Ejsing, C.S.; Conzelmann, A. Saccharomyces cerevisiae is dependent on vesicular traffic between the Golgi apparatus and the vacuole when inositolphosphorylceramide synthase Aur1 is inactivated. Eukaryot. Cell 2015, 14, 1203–1216.
- Cerantola, V.; Guillas, I.; Roubaty, C.; Vionnet, C.; Uldry, D.; Knudsen, J.; Conzelmann, A. Aureobasidin A arrests growth of yeast cells through both ceramide intoxication and deprivation of essential inositolphosphorylceramides. Mol. Microbiol. 2009, 71, 1523–1537.
- Epstein, S.; Castillon, G.A.; Qin, Y.; Riezman, H. An essential function of sphingolipids in yeast cell division. Mol. Microbiol. 2012, 84, 1018–1032.
- Esch, B.M.; Limar, S.; Bogdanowski, A.; Gournas, C.; More, T.; Sundag, C.; Walter, S.; Heinisch, J.J.; Ejsing, C.S.; André, B.; et al. Uptake of exogenous serine is important to maintain sphingolipid homeostasis in Saccharomyces cerevisiae. PLoS Genet. 2020, 16, e1008745.
- Han, S.; Lone, M.A.; Schneiter, R.; Chang, A. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc Natl. Acad. Sci. USA 2010, 107, 5851–5856.
- Breslow, D.K.; Collins, S.R.; Bodenmiller, B.; Aebersold, R.; Simons, K.; Shevchenko, A.; Ejsing, C.S.; Weissman, J.S. Orm family proteins mediate sphingolipid homeostasis. Nature 2010, 463, 1048–1053.
- Niles, B.J.; Powers, T. Plasma membrane proteins Slm1 and Slm2 mediate activation of the AGC kinase Ypk1 by TORC2 and sphingolipids in S. cerevisiae. Cell Cycle 2012, 11, 3745–3749.
- Fröhlich, F.; Moreira, K.; Aguilar, P.S.; Hubner, N.C.; Mann, M.; Walter, P.; Walther, T.C. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J. Cell Biol. 2009, 185, 1227–1242.
- Roelants, F.M.; Breslow, D.K.; Muir, A.; Weissman, J.S.; Thorner, J. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2011, 108, 19222–19227.
- Schmidt, O.; Weyer, Y.; Baumann, V.; Widerin, M.A.; Eising, S.; Angelova, M.; Schleiffer, A.; Kremser, L.; Lindner, H.; Peter, M.; et al. Endosome and Golgi-associated degradation (EGAD) of membrane proteins regulates sphingolipid metabolism. EMBO J. 2019, 38, e101433.
- Muir, A.; Ramachandran, S.; Roelants, F.M.; Timmons, G.; Thorner, J. TORC2-dependent protein kinase Ypk1 phosphorylates ceramide synthase to stimulate synthesis of complex sphingolipids. eLife 2014, 3, e03779.
- Aronova, S.; Wedaman, K.; Aronov, P.A.; Fontes, K.; Ramos, K.; Hammock, B.D.; Powers, T. Regulation of ceramide biosynthesis by TOR complex 2. Cell Metab. 2008, 7, 148–158.
- Fröhlich, F.; Olson, D.K.; Christiano, R.; Farese, R.V., Jr.; Walther, T.C. Proteomic and phosphoproteomic analyses of yeast reveal the global cellular response to sphingolipid depletion. Proteomics 2016, 16, 2759–2763.
- Chauhan, N.; Han, G.; Somashekarappa, N.; Gable, K.; Dunn, T.; Kohlwein, S.D. Regulation of sphingolipid biosynthesis by the morphogenesis checkpoint kinase Swe1. J. Biol. Chem. 2016, 291, 2524–2534.
- Chauhan, N.; Visram, M.; Cristobal-Sarramian, A.; Sarkleti, F.; Kohlwein, S.D. Morphogenesis checkpoint kinase Swe1 is the executor of lipolysis-dependent cell-cycle progression. Proc Natl. Acad. Sci. USA 2015, 112, E1077–E1085.
- Meggio, F.; Pinna, L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003, 17, 349–368.
- Trembley, J.H.; Wang, G.; Unger, G.; Slaton, J.; Ahmed, K. Protein kinase CK2 in health and disease. Cell. Mol. Life Sci. 2009, 66, 1858–1867.
- Fresques, T.; Niles, B.; Aronova, S.; Mogri, H.; Rakhshandehroo, T.; Powers, T. Regulation of ceramide synthase by casein kinase 2-dependent phosphorylation in Saccharomyces cerevisiae. J. Biol. Chem. 2015, 290, 1395–1403.
- González, A.; Hall, M.N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017, 36, 397–408.
- Yan, G.; Shen, X.; Jiang, Y. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J. 2006, 25, 3546–3555.
- Zimmermann, C.; Santos, A.; Gable, K.; Epstein, S.; Gururaj, C.; Chymkowitch, P.; Pultz, D.; Rødkær, S.V.; Clay, L.; Bjørås, M.; et al. TORC1 inhibits GSK3-mediated Elo2 phosphorylation to regulate very long chain fatty acid synthesis and autophagy. Cell Rep. 2013, 5, 1036–1046.
- Olson, D.K.; Fröhlich, F.; Christiano, R.; Hannibal-Bach, H.K.; Ejsing, C.S.; Walther, T.C. Rom2-dependent phosphorylation of Elo2 controls the abundance of very long-chain fatty acids. J. Biol. Chem. 2015, 290, 4238–4247.
- Swinnen, E.; Wilms, T.; Idkowiak-Baldys, J.; Smets, B.; De Snijder, P.; Accardo, S.; Ghillebert, R.; Thevissen, K.; Cammue, B.; De Vos, D.; et al. The protein kinase Sch9 is a key regulator of sphingolipid metabolism in Saccharomyces cerevisiae. Mol. Biol. Cell 2014, 25, 196–211.
- Urban, J.; Soulard, A.; Huber, A.; Lippman, S.; Mukhopadhyay, D.; Deloche, O.; Wanke, V.; Anrather, D.; Ammerer, G.; Riezman, H.; et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 2007, 26, 663–674.
- Loewith, R. TORC1 signaling in budding yeast. In The Enzymes; Academic Press: Cambridge, MA, USA, 2010; Volume 27, pp. 147–175.
- Takahara, T.; Maeda, T. Transient sequestration of TORC1 into stress granules during heat stress. Mol. Cell 2012, 47, 242–252.
- De Craene, J.O.; Bertazzi, D.L.; Bär, S.; Friant, S. Phosphoinositides, major actors in membrane trafficking and lipid signaling pathways. Int. J. Mol. Sci. 2017, 18, 634.
- Kobayashi, T.; Takematsu, H.; Yamaji, T.; Hiramoto, S.; Kozutsumi, Y. Disturbance of sphingolipid biosynthesis abrogates the signaling of Mss4, phosphatidylinositol-4-phosphate 5-kinase, in yeast. J. Biol. Chem. 2005, 280, 18087–18094.
- Gallego, O.; Betts, M.J.; Gvozdenovic-Jeremic, J.; Maeda, K.; Matetzki, C.; Aguilar-Gurrieri, C.; Beltran-Alvarez, P.; Bonn, S.; Fernández-Tornero, C.; Jensen, L.J.; et al. A systematic screen for protein–lipid interactions in Saccharomyces cerevisiae. Mol. Syst. Biol. 2010, 6, 430.
- Jesch, S.A.; Gaspar, M.L.; Stefan, C.J.; Aregullin, M.A.; Henry, S.A. Interruption of inositol sphingolipid synthesis triggers Stt4p-dependent protein kinase C signaling. J. Biol. Chem. 2010, 285, 41947–41960.
- Desrivieres, S.; Cooke, F.T.; Parker, P.J.; Hall, M.N. MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 15787–15793.
- Audhya, A.; Emr, S.D. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell 2002, 2, 593–605.
- Daquinag, A.; Fadri, M.; Jung, S.Y.; Qin, J.; Kunz, J. The yeast PH domain proteins Slm1 and Slm2 are targets of sphingolipid signaling during the response to heat stress. Mol. Cell. Biol. 2007, 27, 633–650.
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175.
- Dunayevich, P.; Baltanás, R.; Clemente, J.A.; Couto, A.; Sapochnik, D.; Vasen, G.; Colman-Lerner, A. Heat-stress triggers MAPK crosstalk to turn on the hyperosmotic response pathway. Sci. Rep. 2018, 8, 15168.
- Yamaguchi, Y.; Katsuki, Y.; Tanaka, S.; Kawaguchi, R.; Denda, H.; Ikeda, T.; Funato, K.; Tani, M. Protective role of the HOG pathway against the growth defect caused by impaired biosynthesis of complex sphingolipids in yeast Saccharomyces cerevisiae. Mol. Microbiol. 2018, 107, 363–386.
- Tani, M.; Funato, K. Protection mechanisms against aberrant metabolism of sphingolipids in budding yeast. Curr. Genet. 2018, 64, 1021–1028.
- Jenkins, G.M.; Richards, A.; Wahl, T.; Mao, C.; Obeid, L.; Hannun, Y. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 32566–32572.
- Sun, Y.; Miao, Y.; Yamane, Y.; Zhang, C.; Shokat, K.M.; Takematsu, H.; Kozutsumi, Y.; Drubin, D.G. Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol. Biol. Cell 2012, 23, 2388–2398.
- Friant, S.; Lombardi, R.; Schmelzle, T.; Hall, M.N.; Riezman, H. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 2001, 20, 6783–6792.
- Schmidt, O.; Weyer, Y.; Sprenger, S.; Widerin, M.A.; Eising, S.; Baumann, V.; Angelova, M.; Loewith, R.; Stefan, C.J.; Hess, M.W.; et al. TOR complex 2 (TORC2) signaling and the ESCRT machinery cooperate in the protection of plasma membrane integrity in yeast. J. Biol. Chem. 2020, 295, 12028–12044.
- Manford, A.G.; Stefan, C.J.; Yuan, H.L.; MacGurn, J.A.; Emr, S.D. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell 2012, 23, 1129–1140.
- Omnus, D.J.; Manford, A.G.; Bader, J.M.; Emr, S.D.; Stefan, C.J. Phosphoinositide kinase signaling controls ER-PM cross-talk. Mol. Biol. Cell 2016, 27, 1170–1180.
- Qian, T.; Li, C.; He, R.; Wan, C.; Liu, Y.; Yu, H. Calcium-dependent and-independent lipid transfer mediated by tricalbins in yeast. J. Biol. Chem. 2021, 296, 100729.
- Thomas, F.B.; Omnus, D.J.; Bader, J.M.; Chung, G.H.; Kono, N.; Stefan, C.J. Tricalbin proteins regulate plasma membrane phospholipid homeostasis. bioRxiv 2021.
- Su, W.C.; Lin, Y.H.; Pagac, M.; Wang, C.W. Seipin negatively regulates sphingolipid production at the ER–LD contact site. J. Cell Biol. 2019, 218, 3663–3680.
