Being the predominant cause of disability, neurological diseases have received much attention from the global health community. Over a billion people suffer from one of the following neurological disorders: dementia, epilepsy, stroke, migraine, meningitis, Alzheimer’s disease, Parkinson’s disease, multiple sclerosis, amyotrophic lateral sclerosis, Huntington’s disease, prion disease, or brain tumors. The diagnosis and treatment options are limited for many of these diseases. Aptamers, being small and non-immunogenic nucleic acid molecules that are easy to chemically modify, offer potential diagnostic and theragnostic applications to meet these needs.
1. Introduction
Aptamers are single-stranded DNA, RNA, or synthetic XNA molecules that can fold into unique three-dimensional (3D) structures by which they bind their target molecules with high specificity and affinity
[1][2]. An approach to selecting aptamers was achieved in 1990 by three separate groups
[3][4][5]. The method is now known as the Systematic Evolution of Ligands by Exponential Enrichment (SELEX). Since then, many aptamers have been selected for use in areas of basic science and an increasing number of aptamers are under development for applications in therapeutics, diagnostics, and imaging
[6][7][8][9][10].
The SELEX method simulates Darwinian evolution in vitro. It includes a number of selection rounds, and, after each round, exponential amplification of the “fittest” oligonucleotides (oligos) is carried out. One round of traditional SELEX includes three steps in the order (1) incubate the desired target molecule with a pool of oligos with a central 20–60 nucleotide region of randomized sequence surrounded by terminal regions of constant sequence that will be templates for later polymerase chain reactions (PCRs), (2) capture the oligos in the pool that successfully bind the target, and (3) amplify the captured oligos by either PCR or reverse transcription-PCR (rt-PCR), depending on the type of oligo, DNA or RNA respectively. Rounds of counter-selection are also included, in which the nucleic acid pool is passed over an empty supporting matrix, without the desired target or over the supporting matrix with one or more other molecules that might be structurally related to the target molecule. In counter selection, the oligos that do not bind the supporting matrix, or the structurally related molecules are captured and continued through more rounds of SELEX. Counter selection is performed for many purposes, such as to eliminate oligos that interact with (1) the supporting matrix, (2) analogs or molecules that are structurally related to the target, or (3) molecules present at high concentrations in the biological matrix (such as serum or tissue extract) in which the selected aptamer will function. The main goal of counter selection is to increase aptamer specificity for the desired target over other potential molecular competitors. Besides conventional SELEX, recently developed structure-switching SELEX is becoming popular
[11][12][13]. In this approach, oligos are required to change their structures when they interact with the target, in order to dissociate from a complementary capture sequence. This ensures that the selected aptamers change in structure upon ligand binding. Other alternative SELEX methods have been developed including CE-SELEX, Cell-SELEX, and in-vivo SELEX, as summarized in
Table 1. It is quite possible to combine different SELEX procedures to develop even more complex techniques. After aptamers are selected by SELEX, they are further modified to improve their specificities and affinities by a number of approaches that, in toto, can be referred to as maturation.
Table 1. Summary of Alternative SELEX methods.
| SELEX Type |
Features |
Reference |
| PhotoSELEX |
Light sensitive oligonucleotides are excited by UV and covalently link to their target molecules |
[14] |
| Cell-SELEX |
Whole cells are used for the selection of aptamers that bind cell surface targets |
[15] |
| In vivo SELEX |
Aptamers are selected from an oligonucleotide pool in living animals |
[16] |
| In silico SELEX |
Computer programs are used to predict tertiary structure, affinity, and target interaction of aptamer candidates |
[17] |
| CE-SELEX |
Capillary electrophoresis is used to select high-affinity aptamers, which reduces the selection process time from weeks to days |
[18] |
| Spiegelmer Technology |
After selection, aptamers are synthesized as unnatural L-oligonucleotides, which are more stable than D-oligonucleotides |
[19] |
| Structure Switching SELEX |
The nucleic acid pool has a short, unvaried sequence by which all oligonucleotides can be captured on a complementary sequence. The oligonucleotides are released when they switch structures to bind their target molecule. |
[13] |
| Magnetic-assisted Rapid Aptamer Selection (MARAS) |
Magnetic nanoparticle-attached targets are used to capture aptamers in the presence of an externally applied rotating magnetic field with varying frequencies that influence the selected aptamer affinities. |
[20] |
| Artificially Expanded Genetic Information (AEGIS)-SELEX |
The AEGIS-SELEX library is composed of oligonucleotides containing natural and non-natural nucleosides. These libraries have higher sequence diversities than libraries of oligonucleotides containing only natural nucleosides. |
[21] |
| Robotic Assisted-SELEX |
Robotic platforms perform the selection without any manual intervention. It reduces the selection process to less than 2 days |
[22] |
| RAPID-SELEX |
A conventional SELEX protocol, but without amplification. After each round, Kd values are measured, and the enriched aptamers are sent to HTS. |
[23] |
| GO-SELEX |
A conventional SELEX protocol with unbound oligos adsorbed by graphene oxide (GO) |
[24] |
| Sol-gel SELEX |
The desired aptamer target is immobilized on a microfluidic device |
[25] |
| Conditional SELEX |
This method enables the selection of aptamers that only function under the chosen condition such as when they are in the presence of a regulatory molecule |
[26] |
| Tailored SELEX |
The library sequences do not have primer complements and SELEX is performed in the absence of primer complements. To amplify the selected oligonucleotides, the primer complements are ligated with primers. This method prevents the primer complements on the oligonucleotides from being part of the selected aptamer structure that binds to target |
[19] |
| SPR-SELEX |
The desired target is immobilized on an SPR chip and the oligo pool injected on the biosensor chip for aptamer selection. |
[27] |
| Chimeric SELEX |
Two or more libraries are used to isolate functionally different aptamers, which are then fused to create a dual function aptamer. |
[28] |
| FRELEX |
Random 8mers are used to capture the aptamers in Phase I and the target molecule is free in solution during Phase II of selection. This method allows for a true free aptamer selection strategy. |
[29] |
There are several considerations in selecting and maturing aptamers. First, the structures of aptamers and aptamer-target interactions are affected by the temperature and the buffer components, including ions, ionic strength, and pH. Thus, one of the most important points in SELEX design is to mimic the environment (biological matrix or salt/ buffer composition) during selection in which the aptamer molecular target is found and the interaction between aptamer and target will occur. Choosing appropriate buffer compositions and incubation temperatures will ensure the optimal performance of the selected aptamers under the conditions in which they will be applied
[4][30][31][32]. Second, aptamer stability may need to be improved, as DNA and RNA aptamers are potential targets for nuclease attack. This is especially the case for RNA, because of the 2′OH group, which is used by ribonucleases in an electrophilic attack of the phosphate of the nucleic acid backbone for hydrolysis. Thus, RNA is more labile to high temperatures and pH than DNA. Aptamers can be matured by post-selection chemical modifications to overcome this susceptibility. However, such modifications can result in the aptamer losing specificity and/or affinity. Therefore, it is usually better to include modified nucleotides during SELEX. Many possible aptamer modifications are listed in
Table 2.
Table 2. Possible Aptamer Modifications with their Characteristics.
| Feature |
Modification |
Reference |
| increases stability and resistance to 3′ exonuclease |
3′-3′ and 5′-5′ internucleotide linkage |
[33] |
| resistance to 3′ exonuclease |
3′ Biotin Conjugates |
[34] |
| increases nuclease resistance |
2′-fluoro (2′-F) Substitution |
[35] |
| 2′-amino (2′NH2) Substitution |
[35] |
| 2′-O-methly (2′-OMe) Substitution |
[36] |
| Triazole replacement |
[37] |
| L-DNA |
[38] |
| increases DNA nuclease resistance, destabilizes quadruplexes in aptamer structure |
thiophosphoryl modifications |
[39] |
| resistance to renal clearance |
5′-End with Cholesterol |
[40] |
| 5′-End with Dialkyl Lipids |
[41] |
| 5′-End with PEGylation |
[42] |
| improving binding affinity and target selectivity |
base modifications (SOMAmers) |
[43] |
| structure-based modifications |
[44] |
Aptamers can be selected that selectively bind most viruses, cells, bacteria, proteins, toxins, and peptides with high affinities
[45][46][47][48]. They can be used several times without noticeable disruption of activity. In other words, they can be separated from their targets for further use. By contrast, antibodies can only be utilized only a few times before they lose functionality. Nucleic acid aptamers can also be matured to be more stable than antibodies and enzymes, particularly at higher temperatures
[2]. Moreover, once the sequence of an aptamer is known, its chemical synthesis and purification to homogeneity are highly reproducible and inexpensive. These properties give aptamers the potential to substitute for antibodies as components of sensing units
[49]. The flexibility of aptamers’ structures and their ability to “mask” regions of an aptamer sequence with complementary oligos provides options for signaling that allow the incorporation of aptamers into many sensor platforms, which are not readily adaptable to antibodies. These properties motivate aptamer-based biosensor development
[50][51][52].
Developing new tools for diagnosis is one of the most salient areas in the biosensor field, with many investigators seeking to create easy, efficient, and cheaper methods to achieve early diagnosis and precision medicine. The inclusion of aptamers in biosensors (named “aptasensors”) took root in the 1990s. An early example of aptamers as bio-recognition elements is an optical biosensor that used fluorescently labeled aptamers against human neutrophil elastase in a homogenous assay
[53]. Since then, many types of aptasensor designs have been reported, most with aptamers on solid supports, and with electrochemical, optical, mechanical, and acoustic signals
[45][54][55][56]. Some examples are discussed later in this article.
Aptamers have also been applied in therapeutics. Modifications that result in increased half-lives and improved pharmacokinetics make some aptamers good options for clinical application. Compared with antibodies, aptamers are much less immunogenic, and their actions can be inhibited by reverse complementary oligonucleotides (Munzar et al., 2019). These important characteristics recommend aptamers for development as therapeutics. Macugen (pegaptanib sodium), which selectively recognizes vascular endothelial growth factor (VEGF), was the first aptamer to be approved as a therapeutic agent (Tobin, 2006). Since then, several aptamers have reached different stages of clinical trials in which they are being tested for treating medical conditions like small cell lung cancer, von Willebrand factor-related disorder, angiomas, and renal cell carcinomas
[51][57][58][59][60].
Another potential therapeutic application of aptamers involves delivery systems
[61][62]. To achieve efficient results in these systems, aptamers should recognize cell surface proteins in their native forms. Cell-SELEX was developed to select against cell-surface proteins that are frequently difficult to purify in their native forms
[63]. An aptamer that recognizes a cell surface protein can coopt the receptor to internalize a cargo attached to the aptamer. With this method, one can select aptamers to receptors primarily expressed on a single cell type that can be targeted in vivo
[57]. For instance, anti-PSMA (prostate-specific membrane antigen) aptamers were developed for the delivery of siRNAs to tumors in mice or cultured cells
[62][64][65][66].
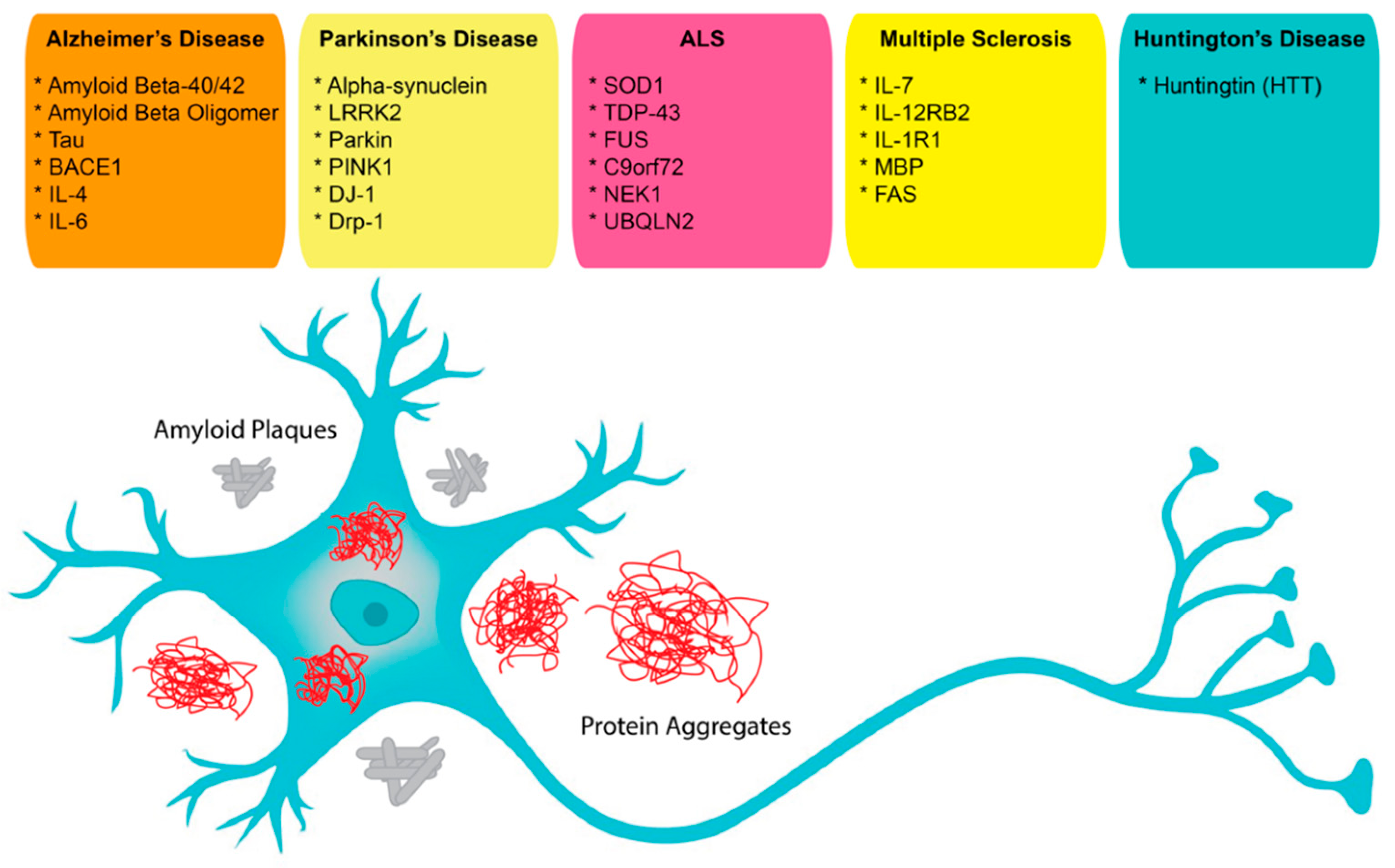
In neuroscience, the use of aptamers has, so far, been limited, albeit while having a very wide range of potential applications. Aptamers have been reported as possible treatment options for neurodegenerative diseases that result in the loss of central nervous system (CNS) structure and function, including Alzheimer’s disease (AD), Parkinson’s disease (PD), Creutzfeldt–Jakob disease, motor neuron diseases, and Huntington’s disease (HD) (
Figure 1). Currently, around 50 million people in the world face dementia, for which the global healthcare cost is close to a trillion US dollars per year. This makes dementias one of the biggest medical and economic problems for our society
[67]. As well, according to the United Nations and the World Health Organization, the world population over 65 years old is estimated to double by 2050. Therefore, age-dependent neurodegenerative diseases will require increasing expenditures for diagnosis and treatment
[67]. These critical demands for methods to facilitate early diagnosis and new therapeutic applications could be met by aptamers, which could also be solutions for some of the underlying complications
[68][69].
This entry is adapted from the peer-reviewed paper 10.3390/ph14121260

