You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Agriculture, Dairy & Animal Science
FN-III proteins are widely distributed in mammals and are usually involved in cellular growth, differentiation, and adhesion. The FNDC5/irisin regulates energy metabolism and is present in different tissues (liver, brain, etc.).
- buffalo
- FN-III proteins
- FNDC5/irisin
- receptors
- binding affinities
1. Introduction
Buffalo (Bubalus bubalis) is a unique livestock species with peculiar productive performance, predominantly found in Asia including China, India, and Pakistan [1,2]. Buffaloes are renowned for their unique ability to consume roughages and convert them into valuable products such as meat and milk. Additionally, buffalo can tolerate harsh weather conditions, perform better under poor feeding resources, and provide draught power [3]. Buffalo milk is relished owing to its peculiar taste with a higher protein, fat, and solid content [4,5,6]. Mainly, buffalo in the Mediterranean and South-Eastern region of Asia serves as an important economic component in the agriculture sector [1,7,8]. Despite having excellent production potential, the productivity of the buffalo is jeopardized due to its poor reproductive efficiency. Buffalo as a dairy animal is known as a poor breeder mainly due to major challenges such as a higher rate of infertility, poor estrus expression [9], poor reproductive efficiency [10], distinct seasonal reproductive pattern [11,12], delayed sexual maturity, prolonged calving intervals [13], and low calf survival rates [14]. It is challenging to improvise the buffalo reproductive and energy metabolism efficiency through finding some biological molecular chaperon that could target reproduction-related signaling receptors to improve the reproductive ability of the buffalo.
In animals, fibronectin proteins are widely dispersed in an extracellular matrix with a variety of functions including cellular growth, migration, differentiation, and adhesion. These proteins are involved in important processes such as healing and the replacement of damaged tissues and embryogenesis [15,16].
In large mammals, the regulation of energy homeostasis under metabolic shifts is the foremost challenge to keep normal physiological and molecular functioning [17]. Fibronectin type III domain containing 5 (FNDC5) was initially designated as a critical factor that causes cellular differentiation of skeletal muscle. Principally, it was detected in peroxisomes [18]. Irisin is a myokine involved in higher energy expenditure through stimulation of white adipose tissues. Firstly, the irisin hormone proteolytically dissociates from its precursor FNDC5, which enhances the circulating irisin levels, subsequently reducing insulin resistance while improving glucose homeostasis [19]. Irisin is mainly secreted from subcutaneous, visceral adipose tissue and skeletal muscles [20,21], but a recent study also reported its presence in other tissues including the spleen, liver, brain, stomach, and testis [22]. The regulatory, molecular, and physiological role of FNDC5/irisin has not yet been fully described and various contradictory findings have been documented in this regard. Thus, there is a dire need to explore the mechanism of FNDC5/irisin functioning in mammals.
2. Identification of FN-III Gene Family and Their Physiochemical Properties
In this study, a comprehensive strategy was applied to characterize the FN-III gene family in the buffalo genome. A total of 29 FN-III genes, widely distributed over different chromosomes of buffalo, harboring variable exons, were detected by using cattle and human as a query sequence and their physiochemical features are presented in Table 1. The FN-III protein isoform’s functional diversity in buffalo was realized from their total number of amino acids ranging from 205 (FNDC5) to 3490 (IGFN1) and MW ranged between 20 kDa and 258 kDa (Table 1). Moreover, according to the instability index, all the members of the FN-III family are unstable except FANK1, LRFN5, IGFN, FLRT1, and FLRT2 which are stable. The isoelectric point indicated that most of the FN-III proteins are acidic (pI < 7), while basic FN-III proteins were also found (pI > 7), as shown in Table 1. Additionally, all of the FN-III proteins have AI values greater than 65 exhibiting thermostable abilities except IGFN1 and FNDC1 having lower AI values (<65), which are seen as thermo-unstable proteins. Furthermore, all of the FN-III proteins behaved as hydrophilic owing to their lower GRAVY values, but FNDC7 and FNDC10 were hydrophobic in nature due to their higher GRAVY values (Table 1).
Table 1. Physiochemical properties of the fibronectin gene family in Bubalus bubalis.
| Gene | Chr. | Exon Count | MW (Da) |
A.A | pI | AI | II | GRAVY |
|---|---|---|---|---|---|---|---|---|
| Fibronectin 1 (FN1) | 2 | 46 | 258,641.53 | 2354 | 5.28 | 69.74 | 40.09 | −0.487 |
| Fibronectin type III domain containing 5 (FNDC5) | 2 | 6 | 22,869.33 | 205 | 6.44 | 92.68 | 52.30 | −0.218 |
| Fibronectin type III domain containing 3B (FNDC3B) | 1 | 31 | 127,736.34 | 1160 | 5.91 | 69.91 | 53.98 | −0.434 |
| Fibronectin type III and ankyrin repeat domains 1 (FANK1) | 23 | 14 | 38,413.93 | 345 | 8.51 | 89.51 | 33.76 | −0.334 |
| Fibronectin type III and SPRY domain containing 1 like (FSD1L) | 3 | 16 | 58,607.09 | 521 | 6.32 | 75.93 | 46.15 | −0.574 |
| Leucine-rich repeat and fibronectin type III domain containing 1 (LRFN1) | 18 | 8 | 82,023.66 | 770 | 7.89 | 90.16 | 49.73 | −0.066 |
| Leucine-rich repeat and fibronectin type III domain containing 5 (LRFN5) | 20 | 8 | 52,122.68 | 466 | 6.60 | 95.47 | 35.44 | −0.141 |
| Fibronectin type III and SPRY domain containing 1 (FSD1) | 9 | 13 | 55,768.58 | 662 | 4.96 | 77.88 | 48.72 | −0.380 |
| Fibronectin type III domain containing 3A (FNDC3A) | 13 | 31 | 133,632.56 | 1217 | 6.44 | 71.27 | 46.88 | −0.412 |
| Fibronectin type III domain containing 1 (FNDC1) | 10 | 23 | 205,865.78 | 1905 | 9.66 | 59.01 | 59.92 | −0.799 |
| Leucine-rich repeat and fibronectin type III domain containing 3 (LRFN3) | 18 | 5 | 72,450.76 | 679 | 9.38 | 87.05 | 59.78 | −0.246 |
| Fibronectin type III and SPRY domain containing 2 (FSD2) | 20 | 15 | 84,755.73 | 747 | 4.81 | 69.69 | 47.20 | −0.593 |
| Fibronectin type III domain containing 7 (FNDC7) | 6 | 13 | 85,949.11 | 811 | 6.53 | 77.69 | 45.18 | 0.046 |
| Ankyrin repeat and fibronectin type III domain containing 1 (ANKFN1) | 3 | 20 | 120,567.79 | 1068 | 6.52 | 80.73 | 58.15 | −0.467 |
| Immunoglobulin like and fibronectin type III domain containing 1 (IGFN1) | 5 | 26 | 347,525.99 | 3490 | 6.49 | 55.35 | 34.98 | −0.590 |
| Fibronectin type III domain containing 4 (FNDC4) | 12 | 7 | 24,753.16 | 230 | 7.66 | 88.87 | 55.08 | −0.252 |
| Fibronectin type III domain containing 8 (FNDC8) | 3 | 4 | 34,298.93 | 312 | 5.29 | 80.93 | 46.44 | −0.370 |
| Leucine-rich repeat and fibronectin type III domain containing 4 (LRFN4) | 5 | 3 | 66,839.10 | 636 | 6.70 | 94.14 | 42.55 | −0.028 |
| Fibronectin type III domain containing protein 3C1-like (LOC102393884) | X | 27 | 157,320.54 | 1433 | 6.79 | 71.84 | 45.92 | −0.439 |
| Fibronectin leucine-rich transmembrane protein 2 (FLRT2) | 11 | 4 | 73,773.40 | 660 | 7.89 | 94.18 | 36.58 | −0.185 |
| EGF like, fibronectin type III and laminin G domains (EGFLAM) | 19 | 23 | 112,751.54 | 1032 | 6.53 | 74.46 | 41.70 | −0.325 |
| Fibronectin type III domain containing 9 (FNDC9) | 9 | 2 | 25,342.98 | 227 | 5.65 | 85.99 | 54.56 | −0.055 |
| Leucine-rich repeat and fibronectin type III domain containing 2 (LRFN2) | 2 | 2 | 87,694.08 | 820 | 6.59 | 90.88 | 44.73 | −0.097 |
| Fibronectin leucine-rich transmembrane protein 3 (FLRT3) | 14 | 3 | 73,171.75 | 649 | 7.56 | 94.18 | 44.53 | −0.296 |
| Fibronectin leucine-rich transmembrane protein 1 (FLRT1) | 5 | 2 | 74,144.68 | 677 | 6.15 | 96.88 | 32.12 | −0.122 |
| Fibronectin type III domain containing 11 (FNDC11) | 14 | 4 | 38,198.37 | 333 | 6.81 | 96.34 | 53.23 | −0.280 |
| Fibronectin type III domain containing 10 (FNDC10) |
5 | 3 | 24,097.32 | 225 | 9.11 | 87.33 | 66.15 | 0.124 |
| Extracellular leucine-rich repeat and fibronectin type III domain containing 2 (ELFN2) | 4 | 4 | 90,363.67 | 824 | 7.78 | 81.78 | 48.76 | −0.295 |
| Extracellular leucine-rich repeat and fibronectin type III domain containing 1 (ELFN1) | 24 | 3 | 87,687.60 | 808 | 8.82 | 79.43 | 61.89 | −0.351 |
[Chr (Chromosome), MW (Molecular Weight in Daltons), A.A (number of amino acids), pI (Isoelectric point), AI (Aliphatic Index), II (Instability Index), and GRAVY (Grand Average of hydropathicity Index)].
3. Gene Structure and Motif Analysis
Furthermore, exon count, phylogenetic relationship, motif pattern, and conserved domain of all the predicted FN-III genes in buffalo were explored (Table 1 and Figure 1). The number of exons varied in all genes; as the highest number of exons were prophesied in FN1, i.e., 46, while the lowest was in FNDC9, i.e., 2 (Table 1). The phylogenetic relationship revealed that all of the buffalo FN-III genes were grouped into nine clades, and ANKFN1, FNDC9, and FANK1 were at the base (Figure 1A). In FN-III genes, ten putative conserved MEME motifs were observed in buffalo (Figure 1B) and five MEME motifs, including motif 1, 2, 4, 6, and 9 corresponding to 50, 50, 41, 50, and 16 amino acids, respectively, were annotated as Leucine-rich repeat domain, while motif 3 and 10 were annotated as fn3 domain after the Pfams search (Table 2). The CDD BLAST was used to confirm the predicted conserved domains in buffalo FN-III genes (Figure 1C). Additionally, the domain of SPRY_PRY_FSD1, BBC, Ig, Glutenin_hmw, PRK04537, PRK07764, PHA03247, Laminin_G_2, LamG, EGF_CA, SPRY, PK12704, DUF5581, DUF4808, DUF5579, PRK15370, PCC, TPKR_C2, Ig_2Ank_2, and ANKYR superfamily has also been dredged up in the buffalo FN-III gene family (Figure 1C).
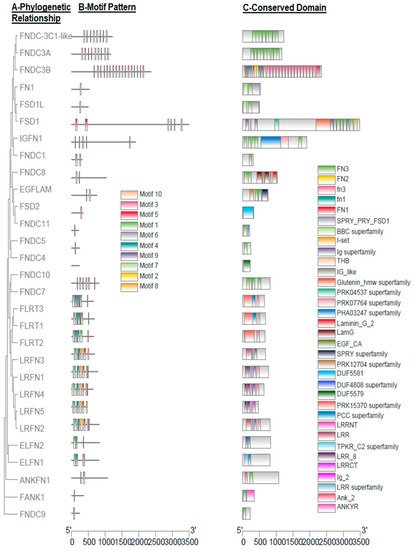
Figure 1. Phylogenetic relationships, motif patterns, and conserved domain regions of buffalo FN-III proteins. (A) Phylogenetic relationship of 29 amino acid sequences of FN-III proteins. (B) Motif pattern. (C) Conserved domain regions. Buffalo ten putative motifs of FN-III proteins are indicated in different colored boxes and details of motifs are enlisted in Table 2.
Table 2. Ten differentially conserved motifs detected in FN-III genes family in Buffalo.
| Motif | Protein Sequence | Length | Pfam Domain |
|---|---|---|---|
| MEME-1 | DNFIAAIPRRDFANMTGLVDLTLSRNTISHIEAGAFDDLENLRALHLDNN | 50 | LRR_8 |
| MEME-2 | NPLHCNCELLWLRRLAREDDLETCASPPGLTGRYFWSVPEEEFLCEPPLI | 50 | LRRCT |
| MEME-3 | LTNLEPDTTYRLCVQALNSAG | 21 | fn3 |
| MEME-4 | MVNLETLRLDHNLIDTIPPGAFSELHKLARLDLTSNRLQKL | 41 | LRR_8 |
| MEME-5 | HWVAPDGRLVGNSSRTRVYPNGTLDILITTSGDSGAFTCIASNAAGEATA | 50 | I-set |
| MEME-6 | CPSVCRCDRGFIYCNDRGLTSIPAGIPEDATTLYLQNNQINNAGIPADLK | 50 | LRRNT |
| MEME-7 | CPKRCICQNLSPSLSTLCAKKGLLFVPPNIDRRTVELRL | 39 | Toxin_11 |
| MEME-8 | WPVQRPAPGIRMYQIQYNSSADDTLVYRM | 29 | - |
| MEME-9 | LEDLDLSYNNLESIPW | 16 | LRR_4 |
| MEME-10 | GTEYRFRVRACNEAGEGPLSEPYTVTTPP | 29 | fn3 |
[LRR_8, Leucine-rich repeat; LRRCT, Leucine-rich repeat C-terminal domain; fn3, Fibronectin type III domain; I-set, Immunoglobulin I-set domain; LRRNT, Leucine-rich repeat N-terminal domain; Toxin_11, Spasmodic peptide gm9a conotoxin from Conus species; LRR_4, Leucine-rich repeats (2 copies)].
4. Collinearity Analysis of FN-III Gene Family
Collinearity analysis showed that genes of the FN-III family in buffalo were distributed over 18 chromosomes, while these genes were present over 21 chromosomes in cattle. Mostly, the buffalo FN-III genes were distributed on proximal or terminal ends of the chromosomes as presented in Figure 2.
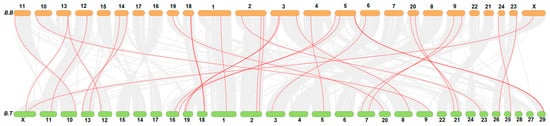
Figure 2. Collinearity analysis of FN-III genes family in buffalo (B.B) and Bos taurus (B.T).
5. Structural Configuration of FNDC5 Protein
For comparative structural configuration, three-dimensional protein models for FNDC5 were also predicted in humans, and different buffalo and cattle breeds (Figure 3). It was observed that FNDC5 protein structures in all species varied with a different number of amino acid residues ranging between 181 and 250. Indeed, there was variation in amino acid residues, but the FNDC5 structure in human, Mediterranean buffalo and cattle was quite similar to each other (Figure 3). Moreover, secondary structural elements including α-helix, β-sheets, transmembrane helix (TM), and degree of disorder also varied in all species. The α-helix was absent in Murrah buffalo and Bos taurus, while human and Mediterranean buffalo breeds shared an approximately similar proportion of β-sheets and TM helix (Table S2). Furthermore, protein in cattle was mainly comprised of α-helix and a higher degree of disorder was observed in buffalo breeds (Table S2).
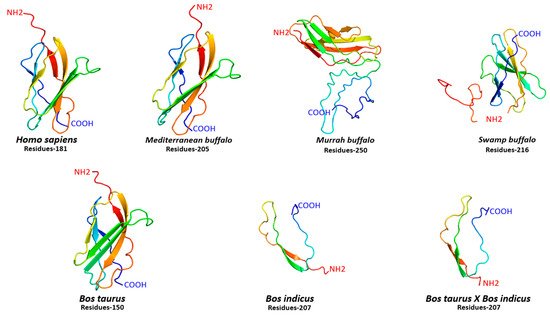
Figure 3. Three-dimensional protein configuration of FNDC5 in humans, cattle, and buffalo.
6. Multiple Sequence Alignment Analysis of Irisin
The comparative amino acid analysis of irisin peptide revealed conserved nature from human to cattle except for Bos indicus and hybrid cattle. Bos indicus and hybrid cattle exhibited a long deletion of 44 amino acids toward the NH2-terminal end. Only a single amino acid variation D106 > G along with 6 amino acid deletion was also observed in Bos taurus at COOH-end. Furthermore, in comparison to humans, all the buffalo breeds had conserved irisin peptide sequences with 100% amino acid sequence homology (Figure 4).

Figure 4. Comparative irisin peptide amino acid analysis of FNDC5 protein in humans, buffalo, and cattle.
7. Molecular Docking Analysis of FNDC5/Irisin
The FNDC5/irisin protein with a molecular weight of 22,869.33 (Dalton) was docked against six receptors to find out the binding affinities. All of the targeted receptors exhibited significant interactions as well as high docking scores ranging from −256.63 to −311.40 (Table 3). A total of 36 hydrogen bonds were detected, which were capable of interacting with the N-terminal portion of all the receptors, except for nuclear receptor subfamily 3 group C member 1 (Table 3, Figure 5 and Figure 6). The FNDC5/irisin also exhibited a strong binding potential with different residues of the selected receptor molecules, where the amino acid residues 36 to 41 were mostly bonded with AR, DCAF6, and ERR-γ (Table 3, Figure 5A,B,D and Figure 6A,B,D). Furthermore, the irisin pocket with amino acid residues ranged between 72 and 91, and showed strong binding potential with ERR-β and KLF15 (Table 3, Figure 5C,E,F and Figure 6C,E,F). The superimposition of FNDC5/irisin (ligand) with all the receptors and their interactions are presented in Figure 5 and Figure 6.
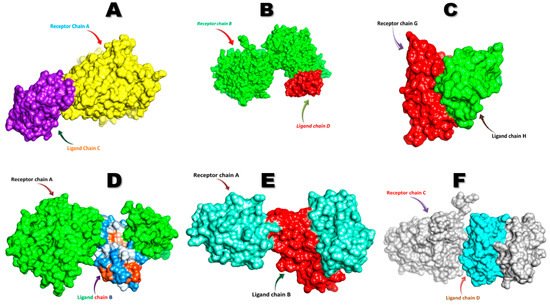
Figure 5. The superimposition of FNDC5 or irisin (ligand) and the receptors (A) Androgen (B) DDB1 and CUL4 associated factor 6 (C) Estrogen-related receptor β (D) Estrogen-related receptor γ (E) Krüppel-like factor 15 (F) Nuclear receptor subfamily 3 group C member 1.
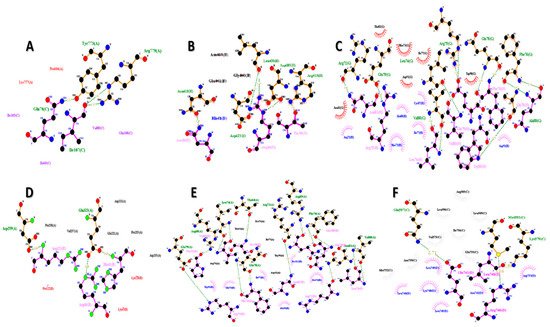
Figure 6. The FNDC5 or irisin (pink; ligand residues) amino acid residues interacting with receptors (A) Androgen (B) DDB1 and CUL4 associated factor 6 (C) Estrogen-related receptor β (D) Estrogen-related receptor γ (E) Krüppel-like factor 15 (F) Nuclear receptor subfamily 3 group C member 1 (all the receptors interacting residues are in green color).
Table 3. Molecular docking results of ligand (FNDC5 or Irisin) binding affinity with different receptors.
| Sr. No. | Receptor | Docking Score |
Ligand RMSD (A0) |
Ligand Interacting Residues |
|---|---|---|---|---|
| 1 | Androgen | −311.40 | 86.42 | Asn36, Thr38, Arg40 |
| 2 | DDB1 and CUL4 associated factor 6 | −256.63 | 79.76 | Asn36, Thr38, Arg40, His41 |
| 3 | Estrogen-related receptor β | −295.57 | 108.96 | Arg72, Mse73, Leu74, Arg75, Phe76, Ile77, Gln78, Glu79, Val80, Asn81, Cys87, Ala88, Trp90, Asp91 |
| 4 | Estrogen-related receptor γ | −256.63 | 79.76 | Arg40, His41, Lys43, Lys120, Pro122, Arg123 |
| 5 | Krüppel-like factor 15 | −260.71 | 81.85 | Ser30, Pro31, Arg72, Mse73, Leu74, Arg75, Phe76, Ile77, Gln78, Glu79, Asn81, Ala88, Trp90, Gln108, Pro112, Val180 |
| 6 | Nuclear receptor subfamily 3 group C member 1 | −308.59 | 108.34 | Lys740, Glu741, Asn742, Leu744, Leu745, Arg746, Leu748, Leu749, Asp753 |
This entry is adapted from the peer-reviewed paper 10.3390/biology10111207
This entry is offline, you can click here to edit this entry!
