Forensic microbiomics is a promising tool for crime investigation. Geolocation connects an individual to a certain place or location by microbiota.
- forensic microbiology
- forensic science
- geolocation
- microbiome
- microbiota
1. Introduction
2. Forensic Microbiome as a Tool for Geolocation
2.1. Soil and Surface Microbiome
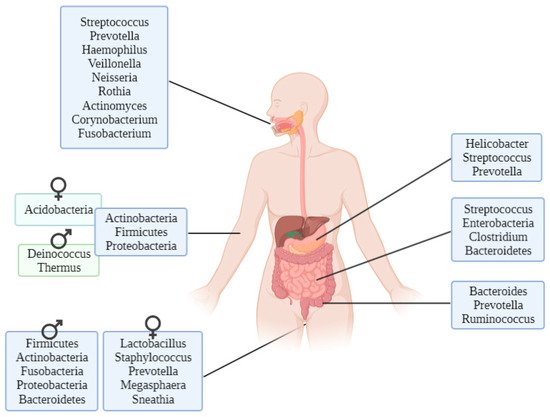
2.2. In Vivo Microbiome
There are genera of microorganisms that allow researchers to assess a person’s geographical origin. For example, Helicobacter pylori extracted from gastric mucosa has been used to determine the geographical origin of unidentified Asian cadavers, resulting in three different clusters: East Asian, Western and Southeast Asian [21]. Furthermore, studies focusing on the relationship between microbiota and diseases such as obesity have found differences between Colombians, Americans, Europeans, Japanese and South Koreans and their relative disposition to increased body mass index [22]. These differences have also been found in studies conducted to evaluate the relationship between the microbiome and infectious diseases such as Plasmodium falciparum infection, finding again geographical differences among people in their stool microbiota [23]. Other studies performed with human hair microbiota have found differences between samples from California and Maryland, and interestingly, scalp hair resulted in better prediction of geolocation than pubic hair [24].
Firmicutes and Bacteroidetes appear to have a certain pattern depending on the latitude. In a study conducted with healthy individuals’ gut microbiota, it was found that the Firmicutes and Bacteroidetes proportion differs with latitude: the proportion of Firmicutes is much higher in the Northern Hemisphere than in the Southern Hemisphere [25]. The explanation of the differences in microbiota remains unclear, although there are three proposed models: host genes, the environment itself or host plasticity.
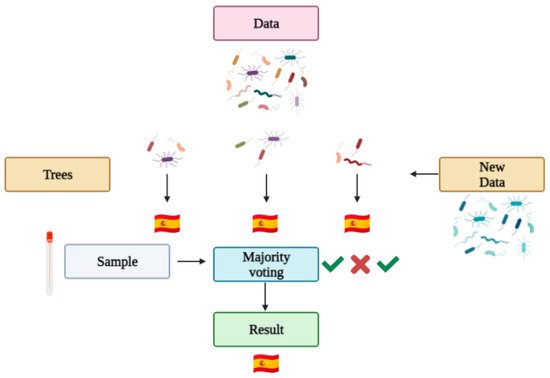
2.3. Machine Learning and Geolocation
2.4. Protocols
2.4.1. Sampling
| Sampling | Samples Should Be Collected Fresh and Then Frozen without Using Any Buffer or Solution. | |
|---|---|---|
| Soil | Swabs | |
| Procedure | Split fresh sample into 2 mL tubes (10) with, at least, 200 mg biomass and store at −80 or −20 °C. | Take 10 replicate swabs with no buffers or solutions and store in −80 or −20 °C |
| Shipping | Samples should be shipped with dry ice in an extruded polystyrene foam container or similar. | |
2.4.2. DNA Extraction
| Commercial Kit | Principle | Format | Time | Automation |
|---|---|---|---|---|
| MagMAX Microbiome Ultra Nucleic Acid Isolation Kit (ThermoFisher Scientific) [37] | Magnetic beads | 100 reactions | ~60 min | KingFisher™ Duo Prime, Flex and Presto |
| Invitrogen PureLink Microbiome DNA Purification Kit (ThermoFisher Scientific) [38] | Spin column | 100 reactions | 120 min | - |
| QIAamp DNA Microbiome Kit (QIAGEN) [39] | Silica columns | 50 reactions | ~180 min | - |
| MO BIO’s PowerMag® Soil DNA Isolation Kit (QIAGEN) [40] | Magnetic beads | 4 × 96 or 32 × 12 | 60–120 min | epMotion® |
2.4.3. Sequencing
3. Challenges and Limitations
This entry is adapted from the peer-reviewed paper 10.3390/life11121322
References
- Kambouris, M.E.; Velegraki, A.; Patrinos, G.P.; Zerva, L. Introduction: The Microbiome as a Concept: Vogue or Necessity? In Microbiomics; Kambouris, M.E., Velegraki, A., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 1–4. ISBN 978-0-12-816664-2.
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103.
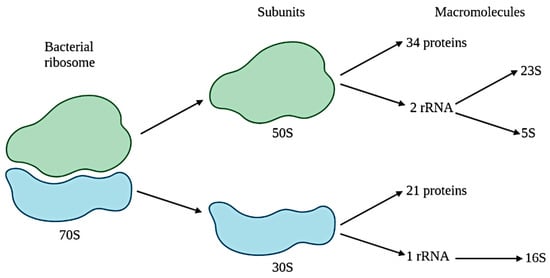
- Fox, G.E.; Magrum, L.J.; Balch, W.E.; Wolfe, R.S.; Woese, C.R. Classification of methanogenic bacteria by 16S ribosomal RNA characterization. Proc. Natl. Acad. Sci. USA 1977, 74, 4537–4541.
- Jan-da, J.M. Taxonomic Classification of Bacteria. In Practical Handbook of Microbiology; Green, L.H., Goldman, E., Eds.; CRC Press: Boca Raton, FL, USA, 2021; pp. 161–167. ISBN 9780367567637.
- Schleifer, K.H. Classification of Bacteria and Archaea: Past, present and future. Syst. Appl. Microbiol. 2009, 32, 533–542.
- Willey, J.M.; Sherwood, L.M.; Woolverton, C.J. Prescott’s Principles of Microbiology; McGraw-Hill: New York, NY, USA, 2009; ISBN 978-0-07-337523-6.
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The Human Microbiome Project. Nature 2007, 449, 804–810.
- Marchesi, J.R. The Human Microbiota and Microbiome; Advances in Molecular and Cellular Microbiology, CABI: Wallingford, UK, 2014; ISBN 9781780640495.
- Budowle, B.; Schutzer, S.E.; Einseln, A.; Kelley, L.C.; Walsh, A.C.; Smith, J.A.L.; Marrone, B.L.; Robertson, J.; Campos, J. Building Microbial Forensics as a Response to Bioterrorism. Science 2003, 301, 1852–1853.
- Bur-cham, Z.M.; Jordan, H.R. History, current, and future use of microorganisms as physical evidence. In Forensic Microbiology; Carter, D.O., Tomberlin, J.K., Benbow, M.E., Metcalf, J.L., Eds.; Wiley: Chichester, UK, 2017; pp. 25–56. ISBN 9781119062554.
- Clarke, T.H.; Gomez, A.; Singh, H.; Nelson, K.E.; Brinkac, L.M. Integrating the microbiome as a resource in the forensics toolkit. Forensic Sci. Int. Genet. 2017, 30, 141–147.
- National Institute of Justice. The Forensic Microbiome: The Invisible Traces We Leave Behind. Available online: https://nij.ojp.gov/topics/articles/forensic-microbiome-invisible-traces-we-leave-behind (accessed on 25 October 2021).
- Robinson, J.M.; Pasternak, Z.; Mason, C.E.; Elhaik, E. Forensic Applications of Microbiomics: A Review. Front. Microbiol. 2021, 11, 3455.
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463.
- Afshinnekoo, E.; Meydan, C.; Chowdhury, S.; Jaroudi, D.; Boyer, C.; Bernstein, N.; Maritz, J.M.; Reeves, D.; Gandara, J.; Chhangawala, S.; et al. Geospatial Resolution of Human and Bacterial Diversity with City-Scale Metagenomics. Cell Syst. 2015, 1, 72–87.
- The MetaSUB International Consortium; Mason, C. The Metagenomics and Metadesign of the Subways and Urban Biomes (MetaSUB) International Consortium inaugural meeting report. Microbiome 2016, 4, 24.
- Danko, D.; Bezdan, D.; Afshinnekoo, E.; Ahsanuddin, S.; Bhattacharya, C.; Butler, D.J.; Chng, K.R.; DeFilippis, F.; Hecht, J.; Kahles, A.; et al. Global Genetic Cartography of Urban Meta genomes and Anti-Microbial Resistance. bioRxiv 2019, 724526.
- Rosenfeld, J.A.; Reeves, D.; Brugler, M.R.; Narechania, A.; Simon, S.; Durrett, R.; Foox, J.; Shianna, K.; Schatz, M.; Gandara, J.; et al. Genome assembly and geospatial phylogenomics of the bed bug Cimex lectularius. Nat. Commun. 2016, 7, 10164.
- Walker, A.R.; Datta, S. Identification of city specific important bacterial signature for the MetaSUB CAMDA challenge microbiome data. Biol. Direct 2019, 14, 11.
- Habtom, H.; Pasternak, Z.; Matan, O.; Azulay, C.; Gafny, R.; Jurkevitch, E. Applying microbial biogeography in soil forensics. Forensic Sci. Int. Genet. 2019, 38, 195–203.
- Escobar, J.S.; Klotz, B.; Valdes, B.E.; Agudelo, G.M. The gut microbiota of Colombians differs from that of Americans, Europeans and Asians. BMC Microbiol. 2014, 14, 311.
- Yooseph, S.; Kirkness, E.F.; Tran, T.M.; Harkins, D.M.; Jones, M.B.; Torralba, M.G.; O’Connell, E.; Nutman, T.B.; Doumbo, S.; Doumbo, O.K.; et al. Stool microbiota composition is associated with the prospective risk of Plasmodium falciparum infection. BMC Genom. 2015, 16, 631.
- Brinkac, L.; Clarke, T.H.; Singh, H.; Greco, C.; Gomez, A.; Torralba, M.G.; Frank, B.; Nelson, K.E. Spatial and Environmental Variation of the Human Hair Microbiota. Sci. Rep. 2018, 8, 9017.
- Suzuki, T.A.; Worobey, M. Geographical variation of human gut microbial composition. Biol. Lett. 2014, 10, 20131037.
- Metcalf, J.L.; Xu, Z.Z.; Bouslimani, A.; Dorrestein, P.; Carter, D.O.; Knight, R. Microbiome Tools for Forensic Science. Trends Biotechnol. 2017, 35, 814–823.
- Knights, D.; Costello, E.K.; Knight, R. Supervised classification of human microbiota. FEMS Microbiol. Rev. 2011, 35, 343–359.
- Pavlov, Y.L. Random Forests; De Gruyter: Berlin, Germany, 2019; Volume 45, ISBN 9783110941975.
- Young, J.M.; Weyrich, L.S.; Breen, J.; Macdonald, L.; Cooper, A. Predicting the origin of soil evidence: High throughput eukaryote sequencing and MIR spectroscopy applied to a crime scene scenario. Forensic Sci. Int. 2015, 251, 22–31.
- Hengl, T.; Nussbaum, M.; Wright, M.N.; Heuvelink, G.B.M.; Gräler, B. Random forest as a generic framework for predictive modeling of spatial and spatio-temporal variables. PeerJ 2018, 6, e5518.
- Grantham, N.S.; Reich, B.J.; Laber, E.B.; Pacifici, K.; Dunn, R.R.; Fierer, N.; Gebert, M.; Allwood, J.S.; Faith, S.A. Global forensic geolocation with deep neural networks. J. R. Stat. Soc. Ser. C Appl. Stat. 2020, 69, 909–929.
- Bjerre, R.D.; Hugerth, L.W.; Boulund, F.; Seifert, M.; Johansen, J.D.; Engstrand, L. Effects of sampling strategy and DNA extraction on human skin microbiome investigations. Sci. Rep. 2019, 9, 17287.
- Thompson, L.; Ackermann, G.; Humphrey, G.; Gilbert, J.; Jansson, J.; Knight, R. EMP Sample Submission Guide v1. protocols.io 2018. Available online: https://www.protocols.io/view/emp-sample-submission-guide-pfqdjmw (accessed on 25 October 2021).
- Marotz, C.; Amir, A.; Humphrey, G.; Gaffney, J.; Gogul, G.; Knight, R. DNA extraction for streamlined metagenomics of diverse environmental samples. Biotechniques 2017, 62, 290–293.
- Minich, J.J.; Zhu, Q.; Janssen, S.; Hendrickson, R.; Amir, A.; Vetter, R.; Hyde, J.; Doty, M.M.; Stillwell, K.; Benardini, J.; et al. KatharoSeq Enables High-Throughput Microbiome Analysis from Low-Biomass Samples. mSystems 2018, 3, e00218-17.
- Alessandrini, F.; Brenciani, A.; Fioriti, S.; Melchionda, F.; Mingoia, M.; Morroni, G.; Tagliabracci, A. Validation of a universal DNA extraction method for human and microbiAL DNA analysis. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 256–258.
- Applied Biosystems Mag MAXTM Microbiome Ultra Nucleic Acid Isolation Kit. Available online: https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2FMAN0018070_MagMAXMicrobiomeNuclAcidIsolatKit_SoilSalivaUrine_Automated_UG.pdf (accessed on 25 October 2021).
- Invitrogen Pure Link TM Microbiome DNA Purification Kit. Available online: https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2FMAN0014331_PureLinkMicrobiome_Soil_UG.pdf (accessed on 25 October 2021).
- QIAGEN QIAamp DNA Microbiome Kit. Available online: https://www.qiagen.com/us/products/discovery-and-translational-research/dna-rna-purification/dna-purification/microbial-dna/qiaamp-dna-microbiome-kit/ (accessed on 25 October 2021).
- QIAGENMO-BIO’s Power Mag Soil DNA Isolation Kit EPH and Book. Available online: https://www.qiagen.com/us/resources/resourcedetail?id=faaba49d-9d10-48cf-a461-01d5302fea49&lang=en (accessed on 25 October 2021).
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4516–4522.
- Amaral-Zettler, L.A.; McCliment, E.A.; Ducklow, H.W.; Huse, S.M. A Method for Studying Protistan Diversity Using Massively Parallel Sequencing of V9 Hypervariable Regions of Small-Subunit Ribosomal RNA Genes. PLoS ONE 2009, 4, e6372.
- European Network of Forensic Science Institutes ENFSI Guideline for Evaluative Reporting in Forensic Science. Available online: https://enfsi.eu/wp-content/uploads/2016/09/m1_guideline.pdf (accessed on 25 October 2021).
- Neckovic, A.; Van Oorschot, R.A.H.; Szkuta, B.; Durdle, A. Challenges in Human Skin Microbial Profiling for Forensic Science: A Review. Genes 2020, 11, 1015.
