Cellular communication plays a critical role in diverse aspects of tumorigenesis including tumor cell growth/death, adhesion/detachment, migration/invasion, angiogenesis, and metastasis. G protein-coupled receptors (GPCRs) which constitute the largest group of cell surface receptors are known to play fundamental roles in all these processes. When considering the importance of GPCRs in tumorigenesis, the adhesion GPCRs (aGPCRs) are unique due to their hybrid structural organization of a long extracellular cell-adhesive domain and a seven-transmembrane signaling domain. Indeed, aGPCRs have been increasingly shown to be associated with tumor development by participating in tumor cell interaction and signaling. ADGRG1/GPR56, a representative tumor-associated aGPCR, is recognized as a potential biomarker/prognostic factor of specific cancer types with both tumor-suppressive and tumor-promoting functions.
- adhesion GPCR
- GPR56
- ligand
- signaling
- tumorigenesis
1. Introduction
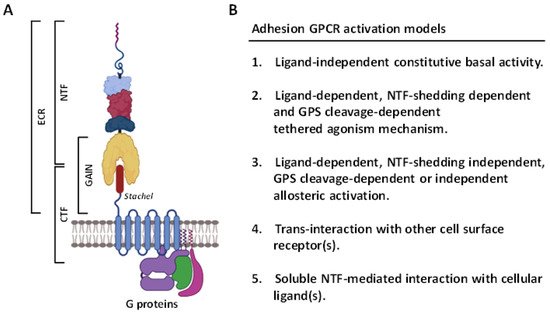
2. Overview of the ADGRG1/GPR56 Receptor
2.1. Structural Characteristics of the ADGRG1/GPR56 Protein
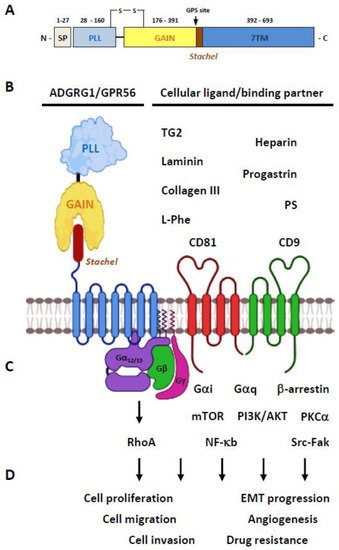
2.2. Ligands/Binding Partners of the ADGRG1/GPR56 Protein
2.3. Activation and Signaling Mechanisms of ADGRG1/GPR56
2.4. The Biological Functions of GPR56
3. ADGRG1/GPR56 as a Cancer Marker and/or Prognostic Factor
| Cancer Type | Function | References |
|---|---|---|
| Melanoma | Potential negative metastatic marker/factor | [28][29][30][83] |
| Acute myeloid leukemia | Leukemia stem cell marker Unfavorable prognostic factor |
[76][81][82][85] |
| Epithelial ovarian cancer | Unfavorable prognostic indicator | [79] |
| Colorectal cancer | Unfavorable prognostic indicator Promote drug-resistant cancer stem cell |
[86][87] |
This entry is adapted from the peer-reviewed paper 10.3390/cells10123352
References
- Marinissen, M.J.; Gutkind, J.S. G-protein-coupled receptors and signaling networks: Emerging paradigms. Trends Pharmacol. Sci. 2001, 22, 368–376.
- Rosenbaum, D.M.; Rasmussen, S.G.; Kobilka, B.K. The structure and function of G-protein-coupled receptors. Nature 2009, 459, 356–363.
- Lefkowitz, R.J. A brief history of G-protein coupled receptors (Nobel Lecture). Angew. Chem. 2013, 52, 6366–6378.
- Bar-Shavit, R.; Maoz, M.; Kancharla, A.; Nag, J.K.; Agranovich, D.; Grisaru-Granovsky, S.; Uziely, B. G protein-coupled receptors in cancer. Int. J. Mol. Sci. 2016, 17, 1320.
- Dorsam, R.T.; Gutkind, J.S. G-protein-coupled receptors and cancer. Nat. Rev. Cancer 2007, 7, 79–94.
- Insel, P.A.; Sriram, K.; Wiley, S.Z.; Wilderman, A.; Katakia, T.; McCann, T.; Yokouchi, H.; Zhang, L.; Corriden, R.; Liu, D.; et al. GPCRomics: GPCR expression in cancer cells and tumors identifies new, potential biomarkers and therapeutic targets. Front. Pharmacol. 2018, 9, 431.
- Gavi, S.; Shumay, E.; Wang, H.Y.; Malbon, C.C. G-protein-coupled receptors and tyrosine kinases: Crossroads in cell signaling and regulation. Trends Endocrinol. Metab. 2006, 17, 48–54.
- Pyne, N.J.; Pyne, S. Receptor tyrosine kinase-G-protein-coupled receptor signalling platforms: Out of the shadow? Trends Pharmacol. Sci. 2011, 32, 443–450.
- Fredriksson, R.; Lagerstrom, M.C.; Lundin, L.G.; Schioth, H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272.
- Hamann, J.; Aust, G.; Arac, D.; Engel, F.B.; Formstone, C.; Fredriksson, R.; Hall, R.A.; Harty, B.L.; Kirchhoff, C.; Knapp, B.; et al. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol. Rev. 2015, 67, 338–367.
- Yona, S.; Lin, H.H.; Siu, W.O.; Gordon, S.; Stacey, M. Adhesion-GPCRs: Emerging roles for novel receptors. Trends Biochem. Sci. 2008, 33, 491–500.
- Arac, D.; Aust, G.; Calebiro, D.; Engel, F.B.; Formstone, C.; Goffinet, A.; Hamann, J.; Kittel, R.J.; Liebscher, I.; Lin, H.H.; et al. Dissecting signaling and functions of adhesion G protein-coupled receptors. Ann. N. Y. Acad. Sci. 2012, 1276, 1–25.
- Arac, D.; Boucard, A.A.; Bolliger, M.F.; Nguyen, J.; Soltis, S.M.; Sudhof, T.C.; Brunger, A.T. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012, 31, 1364–1378.
- Lin, H.H.; Chang, G.W.; Davies, J.Q.; Stacey, M.; Harris, J.; Gordon, S. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J. Biol. Chem. 2004, 279, 31823–31832.
- Paavola, K.J.; Hall, R.A. Adhesion G protein-coupled receptors: Signaling, pharmacology, and mechanisms of activation. Mol. Pharmacol. 2012, 82, 777–783.
- Kishore, A.; Hall, R.A. Versatile signaling activity of adhesion GPCRs. Handb. Exp. Pharmacol. 2016, 234, 127–146.
- Purcell, R.H.; Hall, R.A. Adhesion G protein-coupled receptors as drug targets. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 429–449.
- Stoveken, H.M.; Hajduczok, A.G.; Xu, L.; Tall, G.G. Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc. Natl. Acad. Sci. USA 2015, 112, 6194–6199.
- Liebscher, I.; Schon, J.; Petersen, S.C.; Fischer, L.; Auerbach, N.; Demberg, L.M.; Mogha, A.; Coster, M.; Simon, K.U.; Rothemund, S.; et al. A Tethered Agonist within the Ectodomain Activates the Adhesion G Protein-Coupled Receptors GPR126 and GPR133. Cell Rep. 2015, 10, 1021.
- Monk, K.R.; Hamann, J.; Langenhan, T.; Nijmeijer, S.; Schoneberg, T.; Liebscher, I. Adhesion G protein-coupled receptors: From in vitro pharmacology to in vivo mechanisms. Mol. Pharmacol. 2015, 88, 617–623.
- Piao, X.; Hill, R.S.; Bodell, A.; Chang, B.S.; Basel-Vanagaite, L.; Straussberg, R.; Dobyns, W.B.; Qasrawi, B.; Winter, R.M.; Innes, A.M.; et al. G protein-coupled receptor-dependent development of human frontal cortex. Science 2004, 303, 2033–2036.
- Kim, J.E.; Han, J.M.; Park, C.R.; Shin, K.J.; Ahn, C.; Seong, J.Y.; Hwang, J.I. Splicing variants of the orphan G-protein-coupled receptor GPR56 regulate the activity of transcription factors associated with tumorigenesis. J. Cancer Res. Clin. Oncol. 2010, 136, 47–53.
- Salzman, G.S.; Ackerman, S.D.; Ding, C.; Koide, A.; Leon, K.; Luo, R.; Stoveken, H.M.; Fernandez, C.G.; Tall, G.G.; Piao, X.; et al. Structural basis for regulation of GPR56/ADGRG1 by its alternatively spliced extracellular domains. Neuron 2016, 91, 1292–1304.
- Bae, B.I.; Tietjen, I.; Atabay, K.D.; Evrony, G.D.; Johnson, M.B.; Asare, E.; Wang, P.P.; Murayama, A.Y.; Im, K.; Lisgo, S.N.; et al. Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science 2014, 343, 764–768.
- Knierim, A.B.; Rothe, J.; Cakir, M.V.; Lede, V.; Wilde, C.; Liebscher, I.; Thor, D.; Schoneberg, T. Genetic basis of functional variability in adhesion G protein-coupled receptors. Sci. Rep. 2019, 9, 11036.
- Promel, S.; Langenhan, T.; Arac, D. Matching structure with function: The GAIN domain of adhesion-GPCR and PKD1-like proteins. Trends Pharmacol. Sci. 2013, 34, 470–478.
- Little, K.D.; Hemler, M.E.; Stipp, C.S. Dynamic regulation of a GPCR-tetraspanin-G protein complex on intact cells: Central role of CD81 in facilitating GPR56-Galpha q/11 association. Mol. Biol. Cell 2004, 15, 2375–2387.
- Xu, L.; Begum, S.; Hearn, J.D.; Hynes, R.O. GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc. Natl. Acad. Sci. USA 2006, 103, 9023–9028.
- Yang, L.; Chen, G.; Mohanty, S.; Scott, G.; Fazal, F.; Rahman, A.; Begum, S.; Hynes, R.O.; Xu, L. GPR56 Regulates VEGF production and angiogenesis during melanoma progression. Cancer Res. 2011, 71, 5558–5568.
- Yang, L.; Friedland, S.; Corson, N.; Xu, L. GPR56 inhibits melanoma growth by internalizing and degrading its ligand TG2. Cancer Res. 2014, 74, 1022–1031.
- Yang, L.; Xu, L. GPR56 in cancer progression: Current status and future perspective. Future Oncol. 2012, 8, 431–440.
- Luo, R.; Jeong, S.J.; Jin, Z.; Strokes, N.; Li, S.; Piao, X. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc. Natl. Acad. Sci. USA 2011, 108, 12925–12930.
- Giera, S.; Luo, R.; Ying, Y.; Ackerman, S.D.; Jeong, S.J.; Stoveken, H.M.; Folts, C.J.; Welsh, C.A.; Tall, G.G.; Stevens, B.; et al. Microglial transglutaminase-2 drives myelination and myelin repair via GPR56/ADGRG1 in oligodendrocyte precursor cells. eLife 2018, 7, e33385.
- Luo, R.; Jin, Z.; Deng, Y.; Strokes, N.; Piao, X. Disease-associated mutations prevent GPR56-collagen III interaction. PLoS ONE 2012, 7, e29818.
- Salzman, G.S.; Zhang, S.; Fernandez, C.G.; Arac, D.; Koide, S. Specific and direct modulation of the interaction between adhesion GPCR GPR56/ADGRG1 and tissue transglutaminase 2 using synthetic ligands. Sci. Rep. 2020, 10, 16912.
- Chiang, N.Y.; Chang, G.W.; Huang, Y.S.; Peng, Y.M.; Hsiao, C.C.; Kuo, M.L.; Lin, H.H. Heparin interacts with the adhesion GPCR GPR56, reduces receptor shedding, and promotes cell adhesion and motility. J. Cell Sci. 2016, 129, 2156–2169.
- Jin, G.; Sakitani, K.; Wang, H.; Jin, Y.; Dubeykovskiy, A.; Worthley, D.L.; Tailor, Y.; Wang, T.C. The G-protein coupled receptor 56, expressed in colonic stem and cancer cells, binds progastrin to promote proliferation and carcinogenesis. Oncotarget 2017, 8, 40606–40619.
- Chen, H.; Nwe, P.K.; Yang, Y.; Rosen, C.E.; Bielecka, A.A.; Kuchroo, M.; Cline, G.W.; Kruse, A.C.; Ring, A.M.; Crawford, J.M.; et al. A forward chemical genetic screen reveals gut microbiota metabolites that modulate host physiology. Cell 2019, 177, 1217–1231.
- Li, T.; Chiou, B.; Gilman, C.K.; Luo, R.; Koshi, T.; Yu, D.; Oak, H.C.; Giera, S.; Johnson-Venkatesh, E.; Muthukumar, A.K.; et al. A splicing isoform of GPR56 mediates microglial synaptic refinement via phosphatidylserine binding. EMBO J. 2020, 39, e104136.
- Chang, G.W.; Hsiao, C.C.; Peng, Y.M.; Vieira Braga, F.A.; Kragten, N.A.; Remmerswaal, E.B.; van de Garde, M.D.; Straussberg, R.; Konig, G.M.; Kostenis, E.; et al. The adhesion g protein-coupled receptor GPR56/ADGRG1 is an inhibitory receptor on human NK cells. Cell Rep. 2016, 15, 1757–1770.
- Chiang, N.Y.; Peng, Y.M.; Juang, H.H.; Chen, T.C.; Pan, H.L.; Chang, G.W.; Lin, H.H. GPR56/ADGRG1 activation promotes melanoma cell migration via NTF dissociation and CTF-mediated galpha12/13/RhoA signaling. J. Investig. Dermatol. 2017, 137, 727–736.
- Ohta, S.; Sakaguchi, S.; Kobayashi, Y.; Mizuno, N.; Tago, K.; Itoh, H. Agonistic antibodies reveal the function of GPR56 in human glioma U87-MG cells. Biol. Pharm. Bull. 2015, 38, 594–600.
- Iguchi, T.; Sakata, K.; Yoshizaki, K.; Tago, K.; Mizuno, N.; Itoh, H. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G alpha 12/13 and Rho pathway. J. Biol. Chem. 2008, 283, 14469–14478.
- Salzman, G.S.; Zhang, S.; Gupta, A.; Koide, A.; Koide, S.; Arac, D. Stachel-independent modulation of GPR56/ADGRG1 signaling by synthetic ligands directed to its extracellular region. Proc. Natl. Acad. Sci. USA 2017, 114, 10095–10100.
- Zhu, B.; Luo, R.; Jin, P.; Li, T.; Oak, H.C.; Giera, S.; Monk, K.R.; Lak, P.; Shoichet, B.K.; Piao, X. GAIN domain-mediated cleavage is required for activation of G protein-coupled receptor 56 (GPR56) by its natural ligands and a small-molecule agonist. J. Biol. Chem. 2019, 294, 19246–19254.
- Stoveken, H.M.; Bahr, L.L.; Anders, M.W.; Wojtovich, A.P.; Smrcka, A.V.; Tall, G.G. Dihydromunduletone Is a small-molecule selective adhesion G protein-coupled receptor antagonist. Mol. Pharmacol. 2016, 90, 214–224.
- Stoveken, H.M.; Larsen, S.D.; Smrcka, A.V.; Tall, G.G. Gedunin- and Khivorin-Derivatives are small-molecule partial agonists for adhesion G protein-coupled receptors GPR56/ADGRG1 and GPR114/ADGRG5. Mol. Pharmacol. 2018, 93, 477–488.
- Kishore, A.; Purcell, R.H.; Nassiri-Toosi, Z.; Hall, R.A. Stalk-dependent and stalk-independent signaling by the adhesion G protein-coupled receptors GPR56 (ADGRG1) and BAI1 (ADGRB1). J. Biol. Chem. 2016, 291, 3385–3394.
- Paavola, K.J.; Stephenson, J.R.; Ritter, S.L.; Alter, S.P.; Hall, R.A. The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J. Biol. Chem. 2011, 286, 28914–28921.
- Luo, R.; Jeong, S.J.; Yang, A.; Wen, M.; Saslowsky, D.E.; Lencer, W.I.; Arac, D.; Piao, X. Mechanism for adhesion G protein-coupled receptor GPR56-mediated RhoA activation induced by collagen III stimulation. PLoS ONE 2014, 9, e100043.
- Kishore, A.; Hall, R.A. Disease-associated extracellular loop mutations in the adhesion G protein-coupled receptor G1 (ADGRG1; GPR56) differentially regulate downstream signaling. J. Biol. Chem. 2017, 292, 9711–9720.
- Beliu, G.; Altrichter, S.; Guixa-Gonzalez, R.; Hemberger, M.; Brauer, I.; Dahse, A.K.; Scholz, N.; Wieduwild, R.; Kuhlemann, A.; Batebi, H.; et al. Tethered agonist exposure in intact adhesion/class B2 GPCRs through intrinsic structural flexibility of the GAIN domain. Mol. Cell 2021, 81, 905–921.
- Huang, K.Y.; Lin, H.H. The activation and signaling mechanisms of GPR56/ADGRG1 in Melanoma Cell. Front. Oncol. 2018, 8, 304.
- Moreno, M.; Pedrosa, L.; Pare, L.; Pineda, E.; Bejarano, L.; Martinez, J.; Balasubramaniyan, V.; Ezhilarasan, R.; Kallarackal, N.; Kim, S.H.; et al. GPR56/ADGRG1 inhibits mesenchymal differentiation and radioresistance in glioblastoma. Cell Rep. 2017, 21, 2183–2197.
- Chatterjee, T.; Zhang, S.; Posey, T.A.; Jacob, J.; Wu, L.; Yu, W.; Francisco, L.E.; Liu, Q.J.; Carmon, K.S. Anti-GPR56 monoclonal antibody potentiates GPR56-mediated Src-Fak signaling to modulate cell adhesion. J. Biol. Chem. 2021, 296, 100261.
- Kitakaze, T.; Yoshikawa, M.; Kobayashi, Y.; Kimura, N.; Goshima, N.; Ishikawa, T.; Ogata, Y.; Yamashita, Y.; Ashida, H.; Harada, N.; et al. Extracellular transglutaminase 2 induces myotube hypertrophy through G protein-coupled receptor 56. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118563.
- Della Chiesa, M.; Falco, M.; Parolini, S.; Bellora, F.; Petretto, A.; Romeo, E.; Balsamo, M.; Gambarotti, M.; Scordamaglia, F.; Tabellini, G.; et al. GPR56 as a novel marker identifying the CD56dull CD16+ NK cell subset both in blood stream and in inflamed peripheral tissues. Int. Immunol. 2010, 22, 91–100.
- Peng, Y.M.; van de Garde, M.D.; Cheng, K.F.; Baars, P.A.; Remmerswaal, E.B.; van Lier, R.A.; Mackay, C.R.; Lin, H.H.; Hamann, J. Specific expression of GPR56 by human cytotoxic lymphocytes. J. Leukoc. Biol. 2011, 90, 735–740.
- Ackerman, S.D.; Garcia, C.; Piao, X.; Gutmann, D.H.; Monk, K.R. The adhesion GPCR Gpr56 regulates oligodendrocyte development via interactions with Galpha12/13 and RhoA. Nat. Commun. 2015, 6, 6122.
- Ackerman, S.D.; Luo, R.; Poitelon, Y.; Mogha, A.; Harty, B.L.; D’Rozario, M.; Sanchez, N.E.; Lakkaraju, A.K.K.; Gamble, P.; Li, J.; et al. GPR56/ADGRG1 regulates development and maintenance of peripheral myelin. J. Exp. Med. 2018, 215, 941–961.
- Giera, S.; Deng, Y.; Luo, R.; Ackerman, S.D.; Mogha, A.; Monk, K.R.; Ying, Y.; Jeong, S.J.; Makinodan, M.; Bialas, A.R.; et al. The adhesion G protein-coupled receptor GPR56 is a cell-autonomous regulator of oligodendrocyte development. Nat. Commun. 2015, 6, 6121.
- Jeong, S.J.; Luo, R.; Singer, K.; Giera, S.; Kreidberg, J.; Kiyozumi, D.; Shimono, C.; Sekiguchi, K.; Piao, X. GPR56 functions together with alpha3beta1 integrin in regulating cerebral cortical development. PLoS ONE 2013, 8, e68781.
- Duner, P.; Al-Amily, I.M.; Soni, A.; Asplund, O.; Safi, F.; Storm, P.; Groop, L.; Amisten, S.; Salehi, A. Adhesion G protein-coupled receptor G1 (ADGRG1/GPR56) and pancreatic beta-cell function. J. Clin. Endocrinol. Metab. 2016, 101, 4637–4645.
- Chen, G.; Yang, L.; Begum, S.; Xu, L. GPR56 is essential for testis development and male fertility in mice. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2010, 239, 3358–3367.
- Roly, Z.Y.; Major, A.T.; Fulcher, A.; Estermann, M.A.; Hirst, C.E.; Smith, C.A. Adhesion G-protein-coupled receptor, GPR56, is required for Mullerian duct development in the chick. J. Endocrinol. 2020, 244, 395–413.
- Singh, A.K.; Lin, H.H. The role of GPR56/ADGRG1 in health and disease. Biomed. J. 2021, in press.
- Koirala, S.; Jin, Z.; Piao, X.; Corfas, G. GPR56-regulated granule cell adhesion is essential for rostral cerebellar development. J. Neurosci. 2009, 29, 7439–7449.
- Belzeaux, R.; Gorgievski, V.; Fiori, L.M.; Lopez, J.P.; Grenier, J.; Lin, R.; Nagy, C.; Ibrahim, E.C.; Gascon, E.; Courtet, P.; et al. GPR56/ADGRG1 is associated with response to antidepressant treatment. Nat. Commun. 2020, 11, 1635.
- White, J.P.; Wrann, C.D.; Rao, R.R.; Nair, S.K.; Jedrychowski, M.P.; You, J.S.; Martinez-Redondo, V.; Gygi, S.P.; Ruas, J.L.; Hornberger, T.A.; et al. G protein-coupled receptor 56 regulates mechanical overload-induced muscle hypertrophy. Proc. Natl. Acad. Sci. USA 2014, 111, 15756–15761.
- Wu, M.P.; Doyle, J.R.; Barry, B.; Beauvais, A.; Rozkalne, A.; Piao, X.; Lawlor, M.W.; Kopin, A.S.; Walsh, C.A.; Gussoni, E. G-protein coupled receptor 56 promotes myoblast fusion through serum response factor- and nuclear factor of activated T-cell-mediated signalling but is not essential for muscle development in vivo. FEBS J. 2013, 280, 6097–6113.
- Yeung, J.; Adili, R.; Stringham, E.N.; Luo, R.; Vizurraga, A.; Rosselli-Murai, L.K.; Stoveken, H.M.; Yu, M.; Piao, X.; Holinstat, M.; et al. GPR56/ADGRG1 is a platelet collagen-responsive GPCR and hemostatic sensor of shear force. Proc. Natl. Acad. Sci. USA 2020, 117, 28275–28286.
- Zhang, Y.; Si, Y.; Ma, N.; Mei, J. The RNA-binding protein PCBP2 inhibits Ang II-induced hypertrophy of cardiomyocytes though promoting GPR56 mRNA degeneration. Biochem. Biophys. Res. Commun. 2015, 464, 679–684.
- Maglitto, A.; Mariani, S.A.; de Pater, E.; Rodriguez-Seoane, C.; Vink, C.S.; Piao, X.; Lukke, M.L.; Dzierzak, E. Unexpected redundancy of Gpr56 and Gpr97 during hematopoietic cell development and differentiation. Blood Adv. 2021, 5, 829–842.
- Olaniru, O.E.; Pingitore, A.; Giera, S.; Piao, X.; Castanera Gonzalez, R.; Jones, P.M.; Persaud, S.J. The adhesion receptor GPR56 is activated by extracellular matrix collagen III to improve beta-cell function. Cell. Mol. Life Sci. 2018, 75, 4007–4019.
- Rao, T.N.; Marks-Bluth, J.; Sullivan, J.; Gupta, M.K.; Chandrakanthan, V.; Fitch, S.R.; Ottersbach, K.; Jang, Y.C.; Piao, X.; Kulkarni, R.N.; et al. High-level Gpr56 expression is dispensable for the maintenance and function of hematopoietic stem and progenitor cells in mice. Stem Cell Res. 2015, 14, 307–322.
- Saito, Y.; Kaneda, K.; Suekane, A.; Ichihara, E.; Nakahata, S.; Yamakawa, N.; Nagai, K.; Mizuno, N.; Kogawa, K.; Miura, I.; et al. Maintenance of the hematopoietic stem cell pool in bone marrow niches by EVI1-regulated GPR56. Leukemia 2013, 27, 1637–1649.
- Sud, N.; Sharma, R.; Ray, R.; Chattopadhyay, T.K.; Ralhan, R. Differential expression of G-protein coupled receptor 56 in human esophageal squamous cell carcinoma. Cancer Lett. 2006, 233, 265–270.
- Ke, N.; Sundaram, R.; Liu, G.; Chionis, J.; Fan, W.; Rogers, C.; Awad, T.; Grifman, M.; Yu, D.; Wong-Staal, F.; et al. Orphan G protein-coupled receptor GPR56 plays a role in cell transformation and tumorigenesis involving the cell adhesion pathway. Mol. Cancer Ther. 2007, 6, 1840–1850.
- Liu, Z.; Huang, Z.; Yang, W.; Li, Z.; Xing, S.; Li, H.; Hu, B.; Li, P. Expression of orphan GPR56 correlates with tumor progression in human epithelial ovarian cancer. Neoplasma 2017, 64, 32–39.
- Shashidhar, S.; Lorente, G.; Nagavarapu, U.; Nelson, A.; Kuo, J.; Cummins, J.; Nikolich, K.; Urfer, R.; Foehr, E.D. GPR56 is a GPCR that is overexpressed in gliomas and functions in tumor cell adhesion. Oncogene 2005, 24, 1673–1682.
- Daria, D.; Kirsten, N.; Muranyi, A.; Mulaw, M.; Ihme, S.; Kechter, A.; Hollnagel, M.; Bullinger, L.; Dohner, K.; Dohner, H.; et al. GPR56 contributes to the development of acute myeloid leukemia in mice. Leukemia 2016, 30, 1734–1741.
- Pabst, C.; Bergeron, A.; Lavallee, V.P.; Yeh, J.; Gendron, P.; Norddahl, G.L.; Krosl, J.; Boivin, I.; Deneault, E.; Simard, J.; et al. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood 2016, 127, 2018–2027.
- Liu, M.; Parker, R.M.; Darby, K.; Eyre, H.J.; Copeland, N.G.; Crawford, J.; Gilbert, D.J.; Sutherland, G.R.; Jenkins, N.A.; Herzog, H. GPR56, a novel secretin-like human G-protein-coupled receptor gene. Genomics 1999, 55, 296–305.
- Zendman, A.J.; Cornelissen, I.M.; Weidle, U.H.; Ruiter, D.J.; van Muijen, G.N. TM7XN1, a novel human EGF-TM7-like cDNA, detected with mRNA differential display using human melanoma cell lines with different metastatic potential. FEBS Lett. 1999, 446, 292–298.
- Daga, S.; Rosenberger, A.; Quehenberger, F.; Krisper, N.; Prietl, B.; Reinisch, A.; Zebisch, A.; Sill, H.; Wolfler, A. High GPR56 surface expression correlates with a leukemic stem cell gene signature in CD34-positive AML. Cancer Med. 2019, 8, 1771–1778.
- Lim, D.R.; Kang, D.H.; Kuk, J.C.; Kim, T.H.; Shin, E.J.; Ahn, T.S.; Kim, H.J.; Jeong, D.J.; Baek, M.J.; Kim, N.K. Prognostic impact of GPR56 in patients with colorectal cancer. Neoplasma 2021, 68, 580–589.
- Zhang, S.; Chatterjee, T.; Godoy, C.; Wu, L.; Liu, Q.J.; Carmon, K.S. GPR56 drives colorectal tumor growth and promotes drug resistance through upregulation of MDR1 expression via a RhoA-mediated mechanism. Mol. Cancer Res. 2019, 17, 2196–2207.
