Chloroplasts are plant organelles that carry out photosynthesis, produce various metabolites, and sense changes in the external environment. Given their endosymbiotic origin, chloroplasts have retained independent genomes and gene-expression machinery. Most genes from the prokaryotic ancestors of chloroplasts were transferred into the nucleus over the course of evolution. However, the importance of chloroplast gene expression in environmental stress responses have recently become more apparent. Here, we discuss the emerging roles of the distinct chloroplast gene expression processes in plant responses to environmental stresses. For example, the transcription and translation of psbA play an important role in high-light stress responses. A better understanding of the connection between chloroplast gene expression and environmental stress responses is crucial for breeding stress-tolerant crops better able to cope with the rapidly changing environment.
- chloroplast gene expression
- photosynthesis
- environmental stress response
- transcription
- RNA metabolism
- translation
Chloroplasts are plant organelles that carry out photosynthesis, produce various metabolites, and sense changes in the external environment. Given their endosymbiotic origin, chloroplasts have retained independent genomes and gene-expression machinery. Most genes from the prokaryotic ancestors of chloroplasts were transferred into the nucleus over the course of evolution. However, the importance of chloroplast gene expression in environmental stress responses have recently become more apparent.
- Introduction
Plant often face environmental conditions that are unfavorable for growth and development. These adverse environmental conditions include abiotic and biotic stresses, such as drought, heat, cold, salt, and pathogen infection [1–6]. Environmental stresses pose a great threat to agriculture by limiting crop yields and productivity. The adverse effects of environmental stresses are getting worse due to the increasing worldwide population and climate change. To deal with these environmental stresses, plants rely on their ability to sense and cope with these stresses by regulating the expression of stress-responsive genes in the nucleus, cytoplasm, and organelles.
The chloroplast, a unique plant organelle, is the site of photosynthesis, intracellular signaling, and the production of various compounds important in metabolism, such as amino acids, hormones, nucleotides, vitamins, lipids, and secondary metabolites [7–9]. Chloroplasts also serve as sensors of the external environment. Under stress conditions, chloroplasts send messages to the nucleus through plastid-to-nucleus retrograde signaling, thus optimizing nuclear gene expression based on physiological requirements [7,8]. To date, several possible retrograde signaling pathways have been proposed, including pathways involving intermediates in tetrapyrrole biogenesis [10], the redox state of plastids [11,12], reactive oxygen species [13,14], secondary metabolites in chloroplasts [15,16], and chloroplast gene expression [17–19].
Chloroplasts are semi-autonomous organelles that have retained their own genomes. However, during evolution, most chloroplast genes were lost or transferred to the nucleus: On average, the chloroplast genomes of land plants have retained only 120 genes [20,21]. Nonetheless, these relatively few genes play fundamental roles in chloroplast activities such as energy production and gene expression [22]. Gene expression in chloroplasts is a highly complicated process, far more complex than in their prokaryotic ancestors. This is because chloroplasts have retained a hybrid gene-expression system that combines features of the prokaryotic gene-expression apparatus with eukaryotic innovations (e.g., RNA editing and RNA splicing), and its nascent polycistronic transcripts must undergo many post-transcriptional processing steps [22–25].
The proper expression of chloroplast genes is crucial for chloroplast development and photosynthesis. During the past decade, much effort has been invested in exploring the molecular mechanisms regulating chloroplast gene expression using genetic approaches. Many nucleus-encoded proteins involved in regulating chloroplast gene expression have been identified. However, studies of mutants of these proteins have shown that these mutants are also sensitive to various environmental stresses [8,26–29]. These findings suggest that there is a link between chloroplast gene expression and environmental stress responses, but less attention has been paid to this issue. In this review, we discuss the emerging roles of chloroplast gene expression in plant responses to environmental stresses.
- The Characteristics of Chloroplast Gene Expression
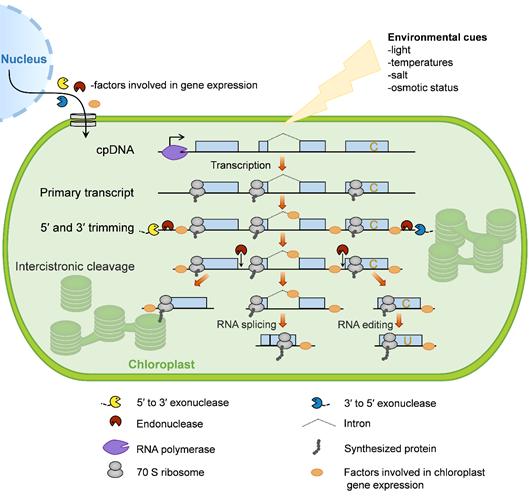
The chloroplast gene-expression system is evolutionarily derived from photosynthetic bacteria that were endocytosed by ancestral eukaryotic plant cells more than 1.5 billion years ago [30]. During evolution, chloroplasts have retained core components of the gene-expression apparatus from their prokaryotic progenitors. In addition, they obtained many eukaryotic properties, such as RNA editing, the prevalence of introns, and complex processing patterns from polycistronic RNA precursors [31]. Here, we briefly describe the processes of chloroplast gene expression in plants (Figure 1).
2.1. Transcription
In plants, chloroplast gene transcription is conducted by two distinct types of RNA polymerases: Nucleus-encoded RNA polymerase (NEP) and plastid-encoded RNA polymerase (PEP) [32,33]. In mature chloroplasts, PEP represents the major transcriptional machinery, which transcribes >80% of all primary chloroplast transcripts, while NEP transcribes chloroplast housekeeping genes [34]. NEP is a phage-type RNA polymerase with a single subunit. In Arabidopsis (Arabidopsis thaliana), NEP is encoded by two nuclear genes, rpoTp and rpoTmp [35]. PEP is a bacteria-type RNA polymerase composed of four core enzyme subunits (α, β, β′, and β′′) and a promoter-recognizing subunit (σ factor). The core enzyme subunits of PEP are encoded by a set of genes located in the plastid genome: rpoA, rpoB, rpoC1, and rpoC2 [33]. By contrast, during evolution, genes for σ factors, which provide the necessary promoter specificity to PEP, were transferred to the nuclear genome, perhaps allowing the nucleus to regulate chloroplast gene transcription in response to environmental and developmental cues [36]. PEP and a set of polymerase-associated proteins (PAPs) form a huge protein complex required for transcription. All PAPs are encoded by genes in the nucleus, and most of them are the components of plastid transcriptionally active chromosome (pTAC) [37]. These PAPs are predicted to be involved in DNA and RNA metabolism (PAP1/pTAC3, PAP2/pTAC2, PAP3/pTAC10, PAP5/pTAC12, PAP7/pTAC14, and PAP12/pTAC7), redox regulation from photosynthesis (PAP6/FLN1, PAP10/TrxZ, and PAP12/pTAC7), and protecting the PEP complex from reactive oxygen species (PAP4/FSD3 and PAP9/FSD2) [38]. The transcriptional regulation of chloroplast genes is essential for the proper functioning of chloroplasts and for overall plant growth under both normal and adverse conditions.
2.2. RNA Metabolism
Most chloroplast genes in plants are organized as operons. These polycistronic primary RNAs require extensive processing, including 5′ and 3′ trimming, intercistronic cleavage, RNA splicing, and RNA editing [39]. Evidence suggests that 5′ and 3′ trimming and intercistronic cleavage are important for moderating RNA stability and translation within chloroplasts [40–44]. In plants, approximately 20 chloroplast genes (encoding proteins or structural RNAs) are interrupted by introns. RNA splicing removes the intron sequences of genes from primary transcripts to enable the production of mature mRNA with the correct genetic information [45,46]. RNA analyses have shown that RNA editing (mainly in the form of C-to-U base conversions) is highly widespread within the chloroplasts of land plants. During this process, numerous C-to-U conversions alter the coding sequences of chloroplast mRNAs, regulate RNA secondary structures that influences the splicing and/or stability of RNAs, or generate translational start sites (AUG) [47,48]. All of these RNA metabolic events depend on many nucleus-encoded proteins, most of which likely arose during coevolution between the host and endosymbiont. For detailed information on chloroplast RNA metabolism, we direct the reader to recent reviews in this area [39,43,45,48,49].
Figure 1. Overview of chloroplast gene expression. In plants, most chloroplast genes are organized as operons and are controlled by single promoters (bent arrow). These genes are transcribed by two distinct types of RNA polymerase: Nucleus-encoded RNA polymerase (NEP) and plastid-encoded RNA polymerase (PEP). The resulting primary transcripts require several processing steps to form mature mRNA, including 5′ and 3′ trimming, intercistronic cleavage, RNA splicing, and RNA editing. In order for these events to take place, numerous nucleus-encoded proteins are translated in the cytosol and imported into the chloroplast, where they control and/or regulate chloroplast gene expression. Chloroplast gene translation is conducted by bacterial-type 70S ribosomes, which occurs cotranscriptionally. Since the mRNA turnover rate within chloroplasts is slow, most ribosomes function in posttranscriptional steps. Moreover, chloroplast gene expression is involved in responses to environmental cues.
2.3. Translation
Chloroplasts possess a bacterial-type 70S ribosome as well as a full set of transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), which conduct protein translation [50]. The 70S ribosome comprises two multi-component subunits: The large (50S) and small (30S) subunits. Both subunits contain rRNAs and various plastid- and nucleus-encoded proteins [51–55].
In general, the chloroplast ribosome has a bacterial-type structure, but with some distinctive features. Chloroplast ribosomes contain the complete set of bacterial-type rRNAs (23S, 16S, and 5S rRNA) with functions analogous to those in bacteria. For example, 23S rRNA exhibits peptidyl transferase activity, whereas 16S rRNA functions as the decoding center and serves as a scaffold for other proteins during ribosome assembly [56,57]. However, the chloroplast contains an additional 4.5S rRNA not found in bacteria that is homologous to the 3′ end of prokaryotic 23S rRNA, suggesting that it was derived from fragmentation of this prokaryotic rRNA [58]. Additionally, two post-transcriptional cleavage sites within the 23S rRNA precursor generate mature 23S rRNA fragments. All of these fragments are assembled into the mature 70S ribosome and combined via intermolecular base pairing [52]. During evolution, obvious changes also occurred in the protein composition of the chloroplast ribosome. The homologs of bacterial proteins Rpl25 and Rpl30 were completely lost in chloroplasts [59]. Several new components of the plastid (chloroplast) ribosome, known as plastid-specific ribosomal proteins (PSRPs), have also been identified [59,60]. PSRP5 and PSRP6 in the 50S subunit and PSRP2 and PSRP3 in the 30S subunit are believed to be intrinsic components of the chloroplast ribosome [52].
- Chloroplast Gene Expression and Environmental Stress
To date, genetic analyses have revealed many nucleus-encoded proteins that regulate not only chloroplast gene expression but also responses to environmental stresses. Functional analyses of these nucleus-encoded proteins have indicated that chloroplast gene expression is involved in plant responses to environmental stresses (Table 1).
3.1. Transcription and Environmental Stress Responses
The transcriptional regulation of chloroplast gene expression is crucial not only for photosynthesis but also for plant development. Recent studies have revealed that the transcriptional control of chloroplast gene expression also plays important roles in plant responses to environmental changes. The chloroplast gene psbA encodes the D1 reaction center protein of photosystem II (PSII) [61–63]. Due to the nature of PSII photochemistry, D1 protein is continuously subjected to photodamage, which decreases photosynthetic activity (an effect known as photoinhibition). These damaged D1 proteins are replaced by de novo synthesized D1 proteins following the partial disassembly of the PSII complex [64,65]. Hence, the capacity to repair photodamaged PSII strongly depends on the ability to generate new D1 protein. Chloroplasts can adjust the transcriptional efficiency of psbA during photoinhibition under adverse environmental conditions such as high light and temperature [66–68]. During chloroplast evolution, several nucleus-encoded proteins have developed the ability to regulate psbA transcription in order to repair photodamaged PSII under adverse environmental conditions. Tomato (Solanum lycopersicum) WHIRLY1 (SlWHY1) was recently found to upregulate psbA transcription under chilling conditions. Under these conditions, the chloroplast-localized SlWHY1 promotes the transcription of psbA by directly binding to the upstream region of its promoter (the sequence “GTTACCCT”), resulting in increased D1 abundance to relieve photoinhibition [69,70]. Overexpression of SlWHY1 leads to increased de novo synthesis of D1 protein and increased resistance to photoinhibition under chilling conditions [69]. These findings suggest that psbA transcription is an important target for regulating PSII activity to adjust plant resistance to environmental stresses.
Table 1. Chloroplast gene expression and stress response mutants.
|
Gene Symbol Accession No. |
Species |
Mutant |
Mutant Stress Phenotype |
Molecular Function |
Reference(s) |
|
Transcription: |
|||||
|
SlWHY1 (Solyc05g007100) |
Solanum lycopersicum |
slwhy1 |
Hypersensitivity to chilling |
Promotes transcription of tomato psbA under chilling conditions |
[69] |
|
SIG5 (AT5G24120) |
Arabidopsis thaliana |
sig5 |
Hypersensitivity to salt stress and high light |
Specifically controls psbD transcription in response to circadian rhythms, environmental stresses, and light signals |
[71] |
|
mTERF5 (AT4G14650) |
Arabidopsis thaliana |
mterf5 |
Decreased sensitivity to salt, ABA, and osmotic stress; altered sugar responses |
Serves as a transcriptional pausing factor; specifically regulates the transcription of chloroplast psbEFLJ |
[72,73] |
|
RNA metabolism: |
|||||
|
ORRM1 (AT3G20930) |
Arabidopsis thaliana |
orrm1 |
Hypersensitivity to low temperature |
Controls RNA editing of 62% (21 of 34) of chloroplast transcripts |
[74,75] |
|
DUA1 (J043016D20) |
Oryza sativa |
dua1 |
Hypersensitivity to low temperature |
Required for RNA editing of the rps8-182 site |
[76] |
|
WSL (Os01g0559500) |
Oryza sativa |
wsl |
Hypersensitivity to ABA, salinity, and sugar with increased H2O2 levels |
Required for splicing of chloroplast rpl2 |
[77] |
|
RH3 (AT5G26742) |
Arabidopsis thaliana |
rh3-4 |
Hypersensitivity to salt stress and low temperature; reduced ABA content |
Involved in splicing of chloroplast trnA, trnI, rpl2, rps12 intron 1, and rps12 intron 2 |
[78–80] |
|
CP29A (AT3G53460) |
Arabidopsis thaliana |
cp29a |
Hypersensitivity to low temperature |
Required for maintaining the stability of various chloroplast transcripts |
[81] |
|
CP31A (AT4G24770) |
Arabidopsis thaliana |
cp31a |
Hypersensitivity to low temperature |
Required for maintaining the stability of various chloroplast transcripts |
[81] |
|
Translation: |
|||||
|
RPS1 (AT5G30510) |
Arabidopsis thaliana |
rps1 |
Heat-sensitive phenotype; perturbed HSF-mediated heat stress response |
Component of chloroplast ribosome small subunit; involved in activating cellular heat stress responses |
[82] |
|
RPS17 (GRMZM2G038013) |
Zea mays |
hcf60-m1 |
Cold-induced bleaching and seedling-lethal phenotype |
Component of chloroplast ribosome small subunit |
[83] |
|
Rps15 (GeneID:800489) |
Nicotiana tabacum |
Δrps15 |
Hypersensitivity to low temperature |
Component of chloroplast ribosome large subunit |
[84] |
|
Rpl33 (GeneID:800444) |
Nicotiana tabacum |
Δrpl33 |
Hypersensitivity to low temperature |
Component of chloroplast ribosome large subunit |
[85] |
The chloroplast gene PsbD encodes the reaction center protein D2 of PSII [86]. The expression of psbD is controlled by four PEP promoters. One of these is the blue-light-responsive promoter psbD BLRP [87–89]. The structure of psbD BLRP is distinct from that of common PEP promoters, which are characterized by conserved –35 and –10 elements. The psbD BLRP contains three cis-elements, including the AAG box, PGT box, and –10 element, but lacks the conserved –35 element [32,89,90]. This promoter has been well characterized. psbD BLRP transcription is specifically regulated by chloroplast-localized sigma factor 5 (SIG5) [71,91]. psbD BLRP transcription is also induced by environmental stresses, such as high salinity, low temperature, and osmotic stress [71]. In addition, psbD BLRP transcription is modulated in response to the relative proportions of red and far red light in a process mediated by signals from phytochromes [92]. Thus, psbD BLRP transcription is modulated during plant responses to environmental stress and sensing of light signals. Indeed, high psbD BLRP activity favors the synthesis of D2, thus relieving high-light-induced damage to PSII [93]. On the other hand, psbD BLRP transcription mediated by SIG5 shows obvious circadian oscillation, revealing how chloroplast gene expression is involved in the circadian oscillator [94].
psbD BLRP transcription may be also involved in biotic stress responses. Pathogens deliver various effectors into plant host cells when pathogens attack plants. These effectors assist pathogen proliferation and suppress plant defense responses [95–101]. Two Pseudomonas effectors, HopR1 and HopBB1, has been suggested to be involved in psbD transcription by targeting PTF1 (PLASTID TRANSCRIPTION FACTOR 1), a transcription factor for psbD BLRP transcription [102,103]. Moreover, the loss of PTF1 leads to more resistant to Pseudomonas syringae pv. tomato strain DC3000 in Arabidopsis [104]. Thus, psbD BLRP transcription may play a role in biotic stress responses.
The psbEFLJ operon contains four chloroplast genes: psbE, psbF, psbL, and psbJ. These genes encode the α and β subunits of cytochrome b559, PsbL, and PsbJ, respectively, which are crucial for the proper functioning of PSII [105,106]. The transcriptional regulation of psbEFLJ was recently investigated. psbEFLJ transcription is positively regulated by the nucleus-encoded protein mTERF5 (mitochondrial Transcription Termination Factor 5), which acts as a pausing factor [72,107]. mTERF5 causes transcriptional pausing on psbEFLJ by binding to the nucleotides +30 to +51 from the transcription start site and recruits additional pTAC6 into the PEP complex at the pausing region to form an enhanced PEP complex, thus positively regulating psbEFLJ transcription. In addition, mterf5 mutants are less sensitive to NaCl and abscisic acid (ABA) than wild-type plants, indicating that mTERF5 functions as a negative regulator of salt tolerance, perhaps via ABA signaling [73]. These findings point to functional links between psbEFLJ transcription and salt tolerance as well as ABA signaling.
3.2. RNA Metabolism and Environmental Stress Responses
RNA metabolism in chloroplasts is remarkably complex, involving a series of steps such as 5′ and 3′ trimming, RNA editing, splicing, and intergenic cleavage [31]. Analyses of mutants with defective RNA editing suggested that RNA editing, splicing, and stability help regulate environmental stress responses in plants [28,29].
An overall deficiency in chloroplast RNA editing (C-to-U base conversion) in Arabidopsis could be caused by the mutation of ORRM1 (Organelle RRM Protein 1), encoding an essential plastid RNA editing factor. orrm1 mutants exhibited greatly reduced RNA editing efficiency compared to wild-type Arabidopsis at 62% (21 of 34) of the chloroplast editing sites. Among these, the editing efficiency at 12 sites decreased by at least 90%, whereas that of the nine other sites decreased by 10% to 90% in orrm1 vs. wild-type plants [74]. The reduced RNA editing deficiency at multiple sites in orrm1 plants did not result in distinctive phenotypes at normal temperatures (22 °C), but the mutants were sensitive to chilling, displaying yellow emerging leaves under chilling conditions (4 °C) [75]. These findings suggest that chloroplast RNA editing confers low-temperature tolerance in Arabidopsis. However, the RNA editing site that confers this improved low-temperature tolerance is unknown.
The indica (Oryza sativa ssp. indica) rice cultivar Dular, referred to as dua1, is planted in tropical regions of Southeast Asia, including India and the Philippines. dua1 plants are less tolerant of low temperatures than Nipponbare (O. sativa ssp. japonica) plants, which are grown in northern areas of Asia, as dua1 plants display pale leaves under low-temperature conditions (19 °C). A recent study revealed that that the low-temperature sensitivity of dua1 is caused by defective RNA editing of the plastid ribosome gene rps8, which is located 182 nt downstream of the translational start site (rps8-182). The edited rps8 transcripts generate RPS8 protein with altered amino acid hydrophobicity, suggesting that RNA editing at rps8-182 improves low-temperature tolerance in rice by moderating the stability of RPS8 protein under low-temperature conditions [76]. Chloroplast genomes have very slow rates of sequence evolution, averaging ~5-fold slower than nuclear genomes [108,109], suggesting that chloroplast RNA editing evolved to improve low-temperature tolerance by increasing protein stability.
ndhB encodes the B subunit of the chloroplast NADH dehydrogenase-like complex that is required for cyclic electron flow around photosystem I [110,111]. The defective RNA editing of ndhB-2, ndhB-3, ndhB-4, and ndhB-6 sites enhances the disease resistance against fungal pathogens in Arabidopsis [112]. This finding suggests that chloroplast RNA editing is interlinked with plant immunity.
rpl2 encodes a component of the 50S subunit in the chloroplast ribosome. This gene contains only a group II intron. In rice, the splicing of this intron is specifically regulated by WHITE STRIPE LEAF (WSL), a pentatricopeptide repeat (PPR) protein. Compared to the wild type, wsl mutants exhibit a decreased germination rate and reduced shoot and root growth upon treatment with ABA but not with α-naphthaleneacetic acid (NAA, an auxin), gibberellic acid (GA), epi-brassinosteroid (BL), or 6-benzylaminopurine (6-BA, a cytokinin). This finding suggests that the ABA signaling process is specifically affected in wsl. These mutants also display decreased germination rates when grown on medium supplemented with sugar and NaCl [77]. Sugar and salinity responses are closely connected with ABA signaling, and several ABA-related genes (e.g., ABI3 and WRKY24) are induced by ABA treatment in wsl mutants, suggesting that rpl2 splicing plays an important role in plant responses to ABA.
In Arabidopsis, the splicing of chloroplast trnA, trnI, rpl2, rps12 intron 1, and rps12 intron 2 is regulated by DEAD-BOX RNA HELICASE 3 (RH3) [78]. Null mutants of RH3 are embryo lethal, whereas the weak allele rh3-4 displays retarded plant growth and pale-green leaves, along with considerable decreases in the splicing efficiency of trnA, trnI, rpl2, rps12 intron 1, and rps12 intron 2. Moreover, the endogenous ABA contents of 1-week-old rh3-4 seedlings are ~50% lower than those of wild-type plants, suggesting that RH3 plays a role in ABA biosynthesis. The mutation of RH3 results in the reduced expression of nucleus-encoded gene ABA1 and NCDE4, encoding two crucial enzymes of the ABA biosynthetic pathway, perhaps explaining the decreased ABA contents of rh3-4 seedlings. Consistent with their decreased ABA contents, rh3-4 mutants exhibit more severely inhibited plant growth and greening than the wild type under abiotic stress conditions including salinity, cold, and dehydration stress [78–80]. These findings suggest that chloroplast RNA splicing of these genes is required for environmental stress responses in plants, especially responses related to ABA signaling. Yet how chloroplast RNA splicing regulates environmental stress responses is currently unknown. A defect in chloroplast RNA splicing would be likely to result in defective photosynthetic performance, thus leading to enhanced sensitivity to environmental stresses. Alternatively, chloroplast RNA splicing might trigger plastid-to-nucleus retrograde signaling to regulate plant stress responses.
Chloroplast RNA stability is also crucial for the proper expression of chloroplast genes. Increasing evidence indicates that chloroplast RNA stability is involved in plant responses to environmental stresses. Chloroplast ribonucleoproteins CP31A and CP29A are RNA chaperone proteins that associate with large sets of chloroplast transcripts [113,81]. Arabidopsis mutants with deletions of CP31A and CP29A do not have unusual phenotypes under normal conditions but show bleaching of newly emerging leaves at the bases of the youngest leaves under cold stress (8 °C). Kupsch et al. demonstrated that CP31A and CP29A are required for the accumulation of transcripts of many chloroplast genes under cold stress (8 °C), such as psaA, psbD, psbF, psbB, petB, ndhF, and rbcL. This cold-sensitive phenotype could be explained by a decreased stability of chloroplast transcripts in the cp31a and cp29a mutants [113,81]. DEAD-box RNA helicase 22 (RH22) is another chloroplast RNA chaperone. In cabbage (Brassica rapa), RH22 expression was significantly upregulated by drought, heat, salt, and cold stress but markedly downregulated by UV stress. The overexpression of cabbage RH22 enhanced the stability of chloroplast transcripts and improved growth and survival in Arabidopsis under drought and salt stress [114]. Moreover, Arabidopsis plants overexpressing cabbage RH22 displayed better growth and more green leaves upon ABA treatment than the wild type, along with decreased expression of ABI3, ABI4, and ABI5, suggesting that chloroplast RNA stability plays a part in ABA signaling pathways [114]. Chloroplast RNA stability might have a positive role in plant responses to environmental stress by enhancing the translation of chloroplast genes.
3.3. Translation and Environmental Stress Responses
Translation is the final step in chloroplast gene expression. Chloroplast gene translation regulates protein accumulation to optimize photosynthetic performance and to attenuate photooxidative damage. Thus, the regulation of chloroplast gene translation represents a unique component of plant responses to internal and external stimuli.
Most plants growing in direct sunlight routinely encounter high-light stress; the resulting high photon flux exceeds the photosynthetic capacity, thereby damaging the chloroplast. To explore the regulation of chloroplast gene translation during the rapid adaptation of plants to high light, a systematic ribosome profiling study was performed to detect changes in chloroplast gene translation efficiency in tobacco seedlings following transfer from moderate light to high light. The ribosome occupancy on psbA transcripts (encoding PSII reaction center protein D1) increased in response to high-light treatment [115]. Given that D1 protein is the main site prone to photodamage by high light, the upregulated psbA translation should substantially facilitate the repair of PSII under high-light stress. However, the molecular mechanisms underlying the translational activation of psbA under these conditions remain to be further explored.
Studies on the functions of chloroplast ribosome proteins have revealed that maintaining sufficiently high chloroplast gene translation efficiency is important for proper chloroplast development at low temperature. Maize (Zea mays) mutants with a loss of ribosomal protein RPS17 were pale green when grown at moderate temperature (27 °C) but appeared albino under cool conditions (17 °C) [83]. Tobacco mutants with a loss of the ribosomal protein Rpl33 showed no visible phenotypes at any stage of development under standard conditions, with similar development, growth rates, and onset of flowering to wild-type plants. However, the Rpl33 knockout mutants were sensitive to cold stress, although not to heat or to low or high light levels. When Rpl33 knockout mutants were transferred to cold-stress conditions (4 °C), they exhibited strong photooxidative damage symptoms and recovered much more slowly from low-temperature stress than wild-type plants [85]. As with Rpl33, the loss of the ribosomal protein Rps15 in tobacco resulted in a growth phenotype almost identical growth to that of wild-type plants, although young plants grew slightly more slowly and the onset of flowering was slightly delayed. However, the Rps15 knockout mutants were cold sensitive, with more severe pigment loss and worse photosynthetic performance than wild-type plants [84]. Together, these findings suggest that the maintenance of plastid translational capacity is important in enabling plant tolerance to chilling stress.
In Arabidopsis, the expression of the chloroplast ribosome protein gene RPS1 was considerably induced by heat stress (2 h at 38 °C). RPS1 knockdown mutants (rps1) displayed retarded growth and slightly pale-green leaves. When rps1 seedlings were exposed to transient high-temperature conditions (3 h at 45 °C), they were much more heat sensitive than wild-type seedlings, as almost no mutants survived after a 7-d recovery, whereas more than 90% of wild-type seedlings did. However, there were no significant differences between rps1 and wild-type plants under osmotic and salinity stress. These results suggest that decreased RPS1 expression alters cellular heat stress responses by disrupting chloroplast gene translation rather than through general physiological defects. RPS1 is required to activate the expression of HsfA2 (HEAT STRESS TRANSCRIPTION FACTOR A-2), a highly heat-shock-inducible gene encoding a transcription factor that is crucial for triggering cellular responses to heat stress. The constitutive expression of HsfA2 was sufficient to rescue the heat-sensitive phenotype of rps1 mutants, suggesting that the defective expression of HsfA2 is responsible for the heat-sensitive phenotype of rps1 mutants. Like the rps1 mutant phenotype, treatment with lincomycin, an inhibitor of chloroplast gene translation, also led to an obvious reduction in the expression of HsfA2 in response to heat stress [82]. These findings reveal a plastid-to-nucleus retrograde signaling pathway that regulates chloroplast gene translational capacity to transcriptionally activate cellular heat stress responses, especially the HsfA2-dependent heat tolerance pathway.
This entry is adapted from the peer-reviewed paper 10.3390/ijms21176082