Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Ecology
Worldwide biodiversity loss points to a necessity of upgrading to a fast and effective monitoring method that can provide quick conservation action. Newly developed environmental DNA (eDNA) based method found to be more cost-effective, non-invasive, quick, and accurate than traditional monitoring (spot identification, camera trapping).
- environmental DNA application
- biodiversity monitoring
- invasive species
1. Introduction
The loss of biodiversity has been one of the most serious concerns worldwide. The world has been losing its biodiversity due to a target to fulfilling high demands of satisfaction by the human race which in turn is incurring expensive and detrimental demands to nature [1]. According to IPBES (The Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services) report, 25% of animals and plants are already threatened with extinction [2]. The wild animals and plants, as well as domestic ones, are facing a fight for survival due to anthropogenic activity. In the next thirty years, 30–50 % of plant species will become extinct [3]. The current rate of extinction is 1000 to 10,000 times greater than the natural extinction rate on our planet [2]. This is an extremely serious issue that will be more severe in the coming days. However, to mitigate this issue, we need to initiate a monitoring program at both local and global levels. In designing such a monitoring system, we need to consider the development of a fit-for-purpose, accurate and cost-effective technique for the detection of species, assessment of biodiversity and study of species interactions.
Environmental DNA, known as eDNA, is shed by organisms during their existence in nature [4]. During their lifespan, organism shed DNA wherever they have been present for a moments. The collection and analysis of these environmental samples and monitoring of the ecosystem without harming organisms is the basis of eDNA study. Recently eDNA, provided valuable contribution to both aquatic and terrestrial monitoring [5,6]. Originally, the eDNA-based species detection was a microbiological study, dating back in 1987 [7] and the use of eDNA to detect macro-organism directly from water sample came to the front in early 2008 with detection of aquatic invasive species [8]. Later on, the methodology was updated and reinforced by some pioneer studies [9,10]. Afterward, rate of eDNA release, degradation, persistence as well as the changes in concentration with organism abundance were explored [11,12,13,14,15,16]. However, as more studies incorporated the use of eDNA approaches, terminology quickly diverged, becoming more convoluted or generally misunderstood [17,18]. What quickly followed was two distinct schools of thought: (i) those who view eDNA as relating to any DNA originating from environmental samples (eDNA sensu lato: [4]), and (ii) research referring to eDNA originating from macro-organisms specifically (eDNA sensu stricto). Researchers use eDNA for species detection to reveal many critical ecological questions, such as studies of population genetics, abundance and habitat preference, detection of unrecorded populations, understanding behavioral biology, monitoring of reproductive migration, pathogens, terrestrial plant community, biodiversity of marine and river ecosystem, nutrient quality, assessment of coral ecosystem, etc. [19]. Moreover, the novel eDNA-based approaches have been used to solve some critical conservation issues such as the detection of rare and endangered species [20], invasive species [21], monitoring whole biodiversity [22,23], study of anthropogenic effect [24], ecosystem health [25] and disease [26]. As eDNA-based methods are emerging rapidly as a multidisciplinary branch of science (Figure 1), it is necessary to evaluate the recent advancements for proper implementation.
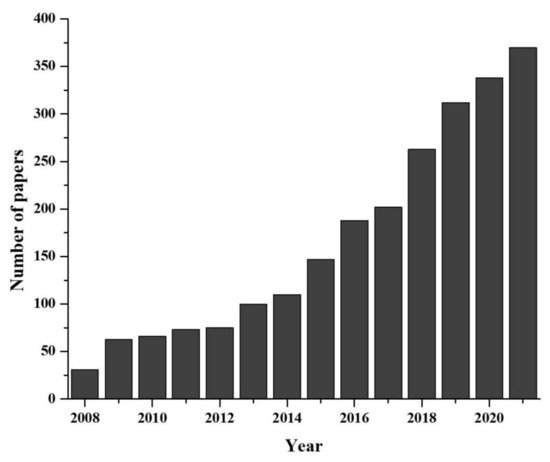
Figure 1. Developmental progress of eDNA technique in last two decades (data collected from PUBMED advanced search with “environmental DNA or eDNA” as title).
This entry is adapted from the peer-reviewed paper 10.3390/biology10121223
This entry is offline, you can click here to edit this entry!
