Cleft lip with or without cleft palate is one of the most common congenital birth defects. This entry aims to identify novel pathogenic microRNAs associated with cleft palate. overexpression of the miR-449 family (miR-449a-3p, miR-449a-5p, miR-449b, miR-449c-3p, and miR-449c-5p) and miR-486b-5p resulted in reduced cell proliferation in primary mouse embryonic palatal mesenchymal (MEPM) cells. On the other hand, inhibitors of miR-130a-3p and miR-301a-3p significantly reduced cell proliferation in MEPM cells. Notably, a miR-130a-3p mimic could ameliorate the cell proliferation defect induced by dexamethasone (a glucocorticoid known to induce cleft palate). Taken together, our results suggest that miR-130-3p plays a crucial role in dexamethasone-induced cleft palate in mice.
1. Introduction
Cleft lip with/without cleft palate (CL/P) is a relatively common congenital birth defect in humans that affects approximately 1 in 700 newborns worldwide
[1]. The palate is composed of the primary palate, which derives from posterior protrusion of nasal processes, and a pair of secondary palates, derived from the lateral protrusion of the maxillary processes. The development of the secondary palate in mammals includes palatal shelf growth, elevation of the palatal shelves, fusion between paired palatal shelves, disappearance of the medial epithelial seam, and intramembranous ossification of the palatal processes of the premaxilla and palatine bone
[2]. Mice have been widely used in the study of palate development, since palate formation and the associated molecular mechanisms of mice are similar to that of humans and occur in a short period of time
[3]. In mice, secondary palate development initiates at embryonic day 11.5 (E11.5) with the formation of tissue folds overlying the future palatal shelves within the oral cavity. Cranial neural-crest-derived mesenchymal cells proliferate within the maxillary processes to form the palatal primordium, which further enlarges to develop the palatal shelves. The palatal shelves continuously grow vertically along the sides of the tongue by E13.5 and then, approximately at E14.0, they elevate to a horizontal position above the tongue. At E14.5, the two palatal shelves meet and start to fuse each at the middle of the oral cavity. Finally, the medial epithelial seam disintegrates by either apoptosis, migration toward epithelial triangles at both oral and nasal sides, or epithelial–mesenchymal transition to complete palatal fusion by E16.5. Any failure of these processes leads to CP
[3].
The etiology of CL/P is complicated, with both genetic and environmental factors involved as well as their interactions
[4][5]. As for environmental factors, maternal exposure to smoking and alcohol consumption are known risk factors for CL/P
[3]. In addition, several teratogens (e.g., phenytoin and toxins such as dioxins and heavy metals) are known to cause CP
[4]. Environmental factors are thought to influence expression of non-coding RNAs including microRNAs (miRNAs), which are small RNAs with 21–25 nucleotides that regulate the expression of target genes at post-transcriptional level
[6]. A number of miRNAs have been found in various species to play roles in a wide array of cellular functions during embryonic development, including palate development
[7][8]. We have previously reported that overexpression of miR-374a, miR-133b, and miR-4680-3p inhibits cell proliferation in cultured human palatal mesenchymal cells
[9]. In addition, exposure to
all-trans retinoic acid (
atRA) alters the expression of miR-106-5p
[10] and miR-124-3p
[11] in mouse embryonic palatal shelves. A growing amount of evidence shows that miRNAs play crucial roles in development and pathological conditions; therefore, it is important to identify how the expression of miRNAs is altered under specific conditions and in presence of chemicals known to cause of CP.
Dexamethasone (DEX) is a synthetic glucocorticoid (GC) clinically used for its anti-inflammatory and immunosuppressive actions through interference with various signaling pathways and molecules, including Toll-like receptors and mitogen-activated protein kinases
[12]. GC signaling acts as either a transactivator or transrepressor of the target genes under physiological and pathological conditions. GCs in the extracellular fluid diffuse into the cytosol and bind to the GC receptor (GR) in the cytosol. In absence of GCs, nuclear protein GR forms a complex with heat shock protein 70 (HSP70), HSP90, FKBP52, and p23 in the cytosol. In presence of GCs, the GC–GR complex releases HSP70/HSP90/FKB92/p23 and forms a dimer of the GC–GR complex. The activated GC–GR dimer translocates into the nucleus and binds to the glucocorticoid response element (GRE) on the promoter region of its target genes, resulting in the activation of transcription (so called transactivation). In addition, the activated GC–GR complex binds to NFκB (p50/p65) without forming a dimer. This NFκB-conjoined monomeric complex represses transcription by binding to the NFκB response element instead of GRE. Thus, gene expression is suppressed (so called transrepression)
[13][14]. Although GCs have tremendous therapeutic usefulness, they are also known for their teratogenicity and toxicity; for example, the oral or systemic administration of corticosteroids increases risk of CL/P two- to nine-fold, a risk of preterm birth or low birth weight
[15][16][17], GC-induced osteonecrosis of the femoral head (GIONFH)
[18], and GC-induced osteoporosis (GOI)
[19]. Furthermore, DEX is known to penetrate the blood–placental barrier and bind to GR in the cytoplasm, causing CP in mice due to suppression of cell proliferation in the palatal mesenchyme
[20][21], and craniofacial dysmorphism by upregulated
mmp13 expression in zebrafish
[22][23]. Although GC treatment induces expression of both genes and miRNAs, the regulatory network of genes and miRNAs remains largely unknown.
2. Genes and miRNAs Potentially Involved in Palate Development
Through secondary data analyses of the miRNA-seq and RNA-seq datasets available at FaceBase, we identified a total of nine miRNAs that were differentially expressed in the palate between E13.5 and E14.5, with a false discovery rate (FDR) < 0.05. A total of five miRNAs (miR-449a-3p, miR-449a-5p, miR-449b, miR-449c-3p, and miR-449c-5p) were upregulated, and a total of four miRNAs (miR-19a-3p, miR-130a-3p, miR-301a-3p, and miR-486b-5p) were downregulated at E14.5 compared to E13.5. To identify genes anti-correlated with the expression of these miRNAs, we queried these miRNAs with four different sequence-based target prediction databases: TargetScan, mirDB, miRWalk, and miRTarBase.
3. miRNAs Involved in Cell Growth in MEPM Cells
First, we analyzed the expression of the identified miRNAs in MEPM cells, as well as in the palatal shelves, at E12.5 to E14.5. MiR-130a-3p was highly expressed in MEPM cells, while miR-301a-3p was expressed at moderate level; miR-449c-5p and miR-486b-5p were expressed at lower level, and miR-449c-3p was not detectable. miR-130a-3p was continuously downregulated through E12.5 to E14.5, miR-301a-3p and miR-486b-5p were transiently upregulated at E13.5, miR-449c-3p was detected at E14.5 only, and miR-449c-5p was upregulated at E14.5 compared to E12.5 and E13.5.
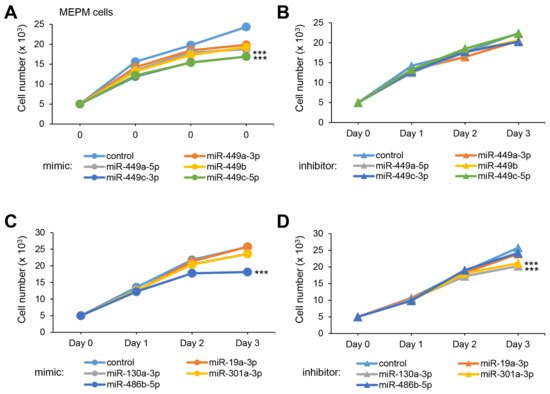
To test whether overexpression or downregulation of these miRNAs could influence cell growth (crucial at E13.5 for the growth of the palatal shelves), we conducted cell growth assays with a mimic and inhibitor for each miRNA and found that all five miR-449 family miRNAs significantly suppressed cell growth in both MEPM and O9-1 cells (Figure 1A). Among them, miR-449c-3p and miR-449c-5p inhibited cell growth more than 30% in both MEPM and O9-1 cells. On the other hand, inhibitors for all of the miR-449 family did not affect cell growth in both MEPM and O9-1 cells (Figure 1B). Moreover, we found that overexpression of miR-486b-5p and suppression of miR-130a-3p and miR-301a-3p inhibited cell growth (Figure 1C,D).
Figure 1. Effect of the predicted miRNAs on cell proliferation in MEPM cells. (A) Cell proliferation assays using MEPM cells from E13.5 palatal shelves treated with the indicated miRNA mimic: control, miR-449a-3p (p < 0.01), miR-449a-5p (p < 0.01), miR-449b (p < 0.01), miR-449c-3p (p < 0.001), and miR-449c-5p mimic (p < 0.001). *** p < 0.001. (B) Cell proliferation assays using MEPM cells treated with control and the indicated miR inhibitor: miR-449a-3p, miR-449a-5p, miR-449b, miR-449c-3p, and miR-449c-5p inhibitor. (C) Cell proliferation assays in MEPM cells treated with the control and the indicated miRNA mimic: miR-19a-3p, miR-130a-3p, miR-301a-3p, and miR-486b-5p (p < 0.001) mimic. *** p < 0.001. (D) Cell proliferation assays in MEPM cells treated with the indicated miRNA inhibitor, control, and miR-19a-3p, miR-130a-3p (p < 0.001), miR-301a-3p (p < 0.001), and miR-486b-5p inhibitor. *** p < 0.001. Each treatment group was compared to the negative control.
To identify genes regulated by either miR-449c-3p, miR-449c-5p, miR-130a-3p, miR-301a-3p, or miR-486b-5p, we conducted qRT-PCR analysis for the predicted genes and found that overexpression of miR-449c-3p significantly downregulated expression of
B43030503Rik,
Cenpb,
Dctn1,
Irf2bpl,
Mink1,
Spint2,
U2af2, and
Zbtb12 in MEPM cells (
Figure 2A). By contrast, suppression of miR-449c-3p significantly upregulated the expression of
B430305J03Rik,
Dctn1,
Spint2,
Srcin1, and
Wnt7b in MEPM cells (
Figure 2C). Overexpression of miR-449c-5p significantly downregulated expression of
Ptms,
Pvrl1, and
Usf1 in MEPM cells (
Figure 2B). Suppression of miR-449c-5p significantly upregulated the expression of
Col17a1,
Pvrl1,
Src, and
Usf1 in MEPM cells (
Figure 2D). Overexpression of miR-130a-3p significantly downregulated expression of
Slc24a2 and
1700028K03Rik in both MEPM cells (
Figure 2E), and suppression of miR-130a-3p significantly upregulated
Slc24a2 expression in MEPM cells (
Figure 2H). Overexpression of miR-301a-3p downregulated
Slc24a2 and
Slc25a46 in MEPM cells (
Figure 2F) and suppression of miR-301a-3p upregulated
Slc24a2 expression in MEPM cells (
Figure 2I). Overexpression of miR-486b-5p significantly downregulated expression of
Fcer1g,
Filip1l,
Kpna2,
Rpl37a, and
Unc5c in MEPM cells (
Figure 2G) and suppression of miR-486b-5p significantly upregulated expression of
Filip1l and
Kpna2 in MEPM cells (
Figure 2J). Taken together, expression of
B430305J03Rik,
Dctn1, and
Spint2 for miR-449c-3p,
Pvrl1 for miR-449c-5p,
Slc24a2 for miR-130a-3p and miR-301a-3p, and
Filip1l for miR-486b-5p was regulated in a dose-dependent manner. Currently, none of these candidate genes have been reported as genes as related to CP
[3]; information available from the CleftGeneDB database (
https://bioinfo.uth.edu/CleftGeneDB, accessed on 27 May 2020). Therefore, we will analyze the function of these candidate genes in cell proliferation.
Figure 2. Effect of the predicted miRNAs on gene expression in MEPM cells. (A) Quantitative RT-PCR for treatment with miR-449c-3p mimic for 24 h in MEPM cells. (B) Quantitative RT-PCR for treatment with miR-449c-5p mimic for 24 h. (C) Quantitative RT-PCR for treatment with miR-449c-3p inhibitor for 24 h. (D) Quantitative RT-PCR for treatment with miR-449c-5p inhibitor for 24 h. (E) Quantitative RT-PCR for treatment with miR-130a-3p mimic for 24 h. (F) Quantitative RT-PCR for treatment with miR-301a-3p mimic for 24 h. (G) Quantitative RT-PCR for treatment with miR-486b-5p mimic for 24 h. (H) Quantitative RT-PCR for the miR-130a-3p inhibitor for 24 h. (I) Quantitative RT-PCR for treatment with miR-301a-3p inhibitor for 24 h. (J) Quantitative RT-PCR for the miR-486b-5p inhibitor treatment for 24 h. * p < 0.05; ** p < 0.01; *** p < 0.001. Each treatment group was compared to the negative control.
4. DEX Suppresses miR-130a-3p Expression in MEPM Cells
Excessive exposure to certain chemicals such as
atRA, phenytoin, and DEX are known to cause CP in mice
[4][24][25]. To investigate whether the expression of candidate miRNAs is associated with chemical exposure, we conducted cell growth assays in MEPM and O9-1 cells under treatment with either DEX,
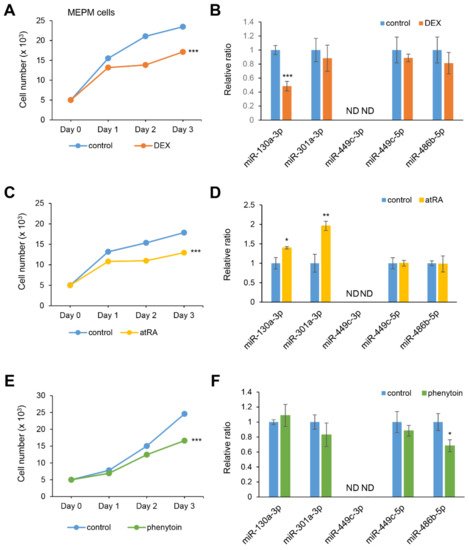
atRA, phenytoin, or vehicle. All three chemicals significantly suppressed cell growth in both MEPM and O9-1 cells (
Figure 3A,C,E). As we expected, miR-130a-3p expression was specifically inhibited under treatment with DEX (
Figure 3B). The expression of miR-130-3p and miR-301a-3p was upregulated under
atRA treatment (
Figure 3D); the expression of miR-486b-5p was significantly suppressed under phenytoin treatment (
Figure 3F). The expression of miR-449c-5p was not altered and miR-449c-3p was not detected under the test conditions. To evaluate the contribution of miR-130a-3p to cell growth under treatment with DEX, MEPM cells were treated with a miR-130a-3p mimic. Notably, the miR-130a-3p mimic completely normalized cell growth under treatment with DEX. The upregulated expression of
Slc24a2, but not
1700028K03Rik, was partially restored by treatment with DEX, in MEPM cells. Thus, our results indicate that DEX can inhibit cell growth through downregulation of miR-130a-3p in MEPM cells.
Figure 3. Influence of DEX, atRA, or phenytoin treatment on cell proliferation and gene expression in MEPM cells. (A,C,E) Cell proliferation assays in MEPM cells treated with 1 µM DEX (A), 10 µM atRA (C), or 50 μg/mL phenytoin (E) for 24, 48, and 72 h. *** p < 0.001 vs. control (n = 6). (B,D,F) Quantitative RT-PCR for the miR-130a-3p, miR-301a-3p, miR-449c-3p, miR-449c-5p, and miR-486b-5p after treatment with DEX (B), atRA (D), or phenytoin (F) for 72 h in MEPM cells. * p < 0.05; ** p < 0.01; *** p < 0.001. Each treatment group was compared to the control. ND; not detectable.
This entry is adapted from the peer-reviewed paper 10.3390/ijms222212453