Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Infectious Diseases
|
Cell Biology
The incidence of sepsis varies depending on the hospital studied, being higher in those dealing with more clinically severe patients, such as cancer hospitals.
- sepsis
- platelets
- microRNA
- membrane antigens
1. Introduction
Early diagnosis and treatment are the main weapons for reducing mortality [1]. The pro-inflammatory response triggered during sepsis leads to activation of the coagulation system and endothelial damage, with the activation of platelets that are stimulated by direct interactions with pathogens [2].
Platelets are small anucleated cells derived from the cytoplasmic fragmentation of bone marrow megakaryocytes. The outer membrane of activated (stimulated) platelets integrates glycoprotein antigens (Gps) and differentiation complexes (CDs), many of which function as receptors and interact with procoagulant factors, adhesion, aggregation, and inhibition molecules. Thus, the increased or decreased expression of these membrane biomarkers is a good indicator of platelet activation [3,4,5].
Thrombocytopenia is commonly associated with sepsis and infections, which in turn are characterized by a profound immune response to the invading pathogen. Platelets exert considerable immunological, antibacterial, and antiviral actions, and therefore are active participants in the host’s response. Platelets contribute to the immune response through several mechanisms, including providing the endothelium with a pro-inflammatory phenotype, increasing and amplifying leukocyte recruitment and inflammation, promoting the effector functions of immune cells, and ensuring an optimal adaptive immune response [6].
Despite the lack of nucleus and genomic DNA, platelets have several types of RNA, ranging from messenger RNAs to non-coding RNAs such as microRNAs (miRNAs), all originating from megakaryocytes. In the activation process, platelets are able to use their own translation mechanism to synthesize proteins, suggesting the possibility of a post-transcriptional gene regulation in these cells [7].
2. Types of Sepsis
Of the 140 patients with sepsis, 18 (12.9%) had uncomplicated sepsis, 22 (15.7%) had severe sepsis, and 100 (71.4%, p < 0.05) developed septic shock. As for the main risk factors, 79 patients with sepsis were older than 65 years (56.4%) and all progressed to death, showing that a higher age favored the susceptibility of these patients.
In total, 110 (78.6%) died regardless of the worsening of the disease. As expected, the greater the aggravation of this pathology, the longer the hospital stay. In addition, the greater the severity of sepsis, the greater the exposure to invasive procedures, thus all patients with septic shock underwent some invasive procedure during their stay in the ICU, mainly through a central vascular catheter, urinary catheter, mechanical ventilation or tracheostomy, which may explain the worsening of sepsis (Table 1).
Table 1. Clinical features of patients admitted to the ICU.
| Variables | n |
|---|---|
| Patients admitted to the ICU | 200 |
| Patients admitted to the ICU with sepsis | 140 (70%) |
| Male gender | 122 (61%) |
| Average age, years (range) | 60.75 (28–89) |
| Average length of stay in the ICU for patients without sepsis (days) | 5 |
| Average length of stay in the ICU for patients with sepsis (days) | 15.7 |
| APACHE II score average | 35 |
| Type of ICU admission—n (%) * | |
| Medical admission | 132 (66%) |
| Surgical admission | 68 (34%) |
| Infection Site—n (%) * | |
| Pulmonary | 91 (65%) |
| Abdominal | 42 (30%) |
| Urinary bacteremia | 7 (5%) |
| Main Comorbidities—n (%) * | |
| Neoplasms | 112 (80%) |
| Diabetes mellitus | 15 (10.7%) |
| Systemic arterial hypertension (PAH) | 13 (9.3%) |
| Invasive Procedures—n (%) *,# | |
| Urinary catheterization | 77 (55%) |
| Central vascular catheterization | 116 (82.9%) |
| Bladder probe | 70 (50%) |
| Tracheostomy | 105 (75%) |
| Mechanical ventilation | 111 (79.3%) |
Legend: * n = 140 (patients with sepsis); # most patients received more than one invasive procedure.
3. Sepsis Etiology
According to the worsening of the disease, bacteria were the main etiological agents (75%), of which Gram-negative bacilli were the most frequent (26%), especially in patients with greater disease severity. From this group, the non-fermenting bacilli (58%) and the Enterobacteriaceae family (52%) stand out. Gram-positive cocci represented 7% of the total of isolated microorganisms, of which 60% were Staphylococcus aureus, 20% Enterococcus sp., and 20% coagulase-negative Staphylococcus. Furthermore, in 67% of cases, other bacteria were also found, such as mycobacteria (69%), Clostridium tetani (19%), and Neisseria meningitidis (12%). Viruses and fungi accounted for 90% of the samples, with COVID-19 being the most frequent (92%), while HIV, H1N1 virus, and Candida albicans yeast were found in 8% of the samples.
Among the microorganisms resistant to the main antimicrobials used in the clinic, Gram-positive bacteria were found in 20% of clinical isolates, of which 50% were Staphylococcus aureus (S. aureus), mainly methicillin-resistant S. aureus (MRSA, 70%). Furthermore, among the coagulase-negative Enterococcus and Staphylococcus (coagulase-negative S.), two clinical isolates of vancomycin-resistant Enterococcus (VRE) and two methicillin-resistant S. coagulase-negative (MRSCON) isolates were identified. Regarding Gram-negative bacteria, 25 clinical isolates were identified, highlighting enterobacteria resistant to third- and fourth-generation cephalosporins (48% of enterobacteria), followed by Pseudomonas aeruginosa (P. aeruginosa) resistant to carbapenems (35% of P. aeruginosa isolates). Acinetobacter baumannii (A. baumannii) resistant to third-generation cephalosporins and carbapenems and Stenotrophomonas maltophilia (S. maltophilia) resistant to sulfamethoxazole and trimethoprim corresponded to 50% and 100% of these species, respectively.
It is noteworthy that 40% (56) of the 140 patients who developed sepsis had the first negative culture results. In these patients, the presence of microorganisms was detected and clinically characterized when sepsis was in full development. Empirical antibiotic treatment was initiated mostly with broad-spectrum drugs such as carbapenems (imipenem, meropenem), third- and fourth-generation cephalosporins, and vancomycin.
4. Platelet Immunophenotyping (Platelet Activation)
The basal level of expression of P2Y12, CD62P, CD41, and CD61 antigens on the surface of platelets showed a significant increase (p < 0.01) between platelets of patients before admission to the ICU and after the development of sepsis (Table 2). This difference was also found in relation to platelets from healthy volunteers (negative control).
Table 2. Parameters evaluated in platelets of patients and volunteers (controls).
| Parameters | Healthy Controls | Before Uncomplicated Sepsis | In Uncomplicated Sepsis | Before Severe Sepsis | Severe Sepsis | Before Septic Shock | Septic Shock |
|---|---|---|---|---|---|---|---|
| Membrane Proteins Expression | |||||||
| P2Y12 | 39.28 ± 1.51 | 38.94 ± 0.80 | 101.24 ± 3.27 * | 40.87 ± 0.89 | 166.64 ± 20.34 * | 42.40 ± 1.94 | 281.85 ± 24.89 * |
| CD62P | 9.22 ± 1.30 | 18.39 ± 2.09 | 72.19 ± 3.36 * | 22.69 ± 3.55 | 121.06 ± 5.30 * | 22.78 ± 3.47 | 159.97 ± 6.27 * |
| CD41 | 98.87 ± 4.30 | 108.83 ± 4.62 | 233.83 ± 4.49 * | 121.12 ± 4.82 | 315.39 ± 4.72 * | 130.62 ± 4.67 | 462.04 ± 7.52 * |
| CD61 | 87.46 ± 2.09 | 91.25 ± 1.33 | 233.83 ± 4.49 * | 94.31 ± 1.72 | 308.56 ± 1.42 * | 95.51 ± 1.37 | 394.95 ± 13.68 * |
| miRNAs Expression | |||||||
| miR-127 | 0.369 ± 0.03 | 0.371 ± 0.004 | 0.404 ± 0.002 | 0.359 ± 0.033 | 0.442 ± 0.024 | 0.454 ± 0.005 | 0.453 ± 0.030 |
| miR-320a | 0.249 ± 0.02 | 0.281 ± 0.007 | 0.606 ± 0.007 | 0.309 ± 0.007 | 0.756 ± 0.024 | 0.343 ± 0.028 | 0.843 ± 0.028 |
Legend: * p < 0.01 when comparing the values of the patients who developed the three different types of sepsis versus the values of the same patients before sepsis and in relation to the values of healthy controls.
5. Real-Time Expression of miRNAs miR-127 and miR-320a in Platelets
In all 100 volunteers who donated healthy platelets, as expected, the highest quantity was that of miR-127 in relation to miR-320a. Only the 140 patients who developed sepsis had the relative quantity of these miRNAs inverted—that is, the quantity of miR-320a was greater than that of miR-127 (Table 2, Figure 1). The relationship between the expression levels of these two miRNAs is shown in the heatmap in Figure 2, made for 20 patients who developed sepsis.
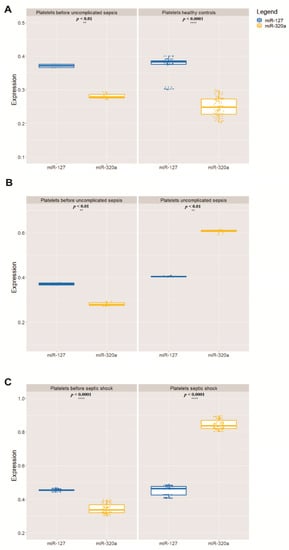
Figure 1. Relative mean expression of miRNAs quantitative PCR (qPCR) analysis (A). The Wilcoxon paired-sample test was applied to estimate the significance of expression of miR-127 and miR-320a in relation to miR-191 (B), which has been used as an internal control for validation analysis (** p < 0.01, **** p < 0.0001) (C). In all graphs, the X-axis represents the storage time of the PCs and the Y-axis represents the 100 PC units.
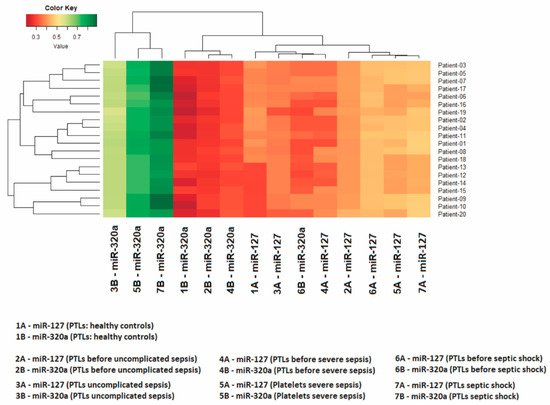
Figure 2. The heatmap shows expression levels in 20 patients. The Z-score was the metric applied to infer the clustering of the heatmap. Gradients with a red tendency represent miRNAs with a lower Z-score and gradients with a green tendency are those with a higher Z-score.
Importantly, the quantitative change in miRNAs was significant (p < 0.01) when comparing the values of patients who developed the three different types of sepsis versus the values of the same patients before sepsis and the values of healthy volunteers. The change in miR-127 and miR-320a also coincided with the development of thrombocytopenia, as the 140 patients who developed sepsis had platelets below 50,000/µL and increased procalcitonin (above 2.0 ng/mL).
This entry is adapted from the peer-reviewed paper 10.3390/genes12121877
This entry is offline, you can click here to edit this entry!
