Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Agriculture, Dairy & Animal Science
Cucumber green mottle mosaic virus (CGMMV), as a typical seed-borne virus, causes costly and devastating diseases in the vegetable trade worldwide.
- seed-borne virus
- cucumber green mottle mosaic virus
- RNA silencing suppressor
1. Introduction
Seed-based transmission of viruses via contaminated seed coats or seed embryos and the infection of germinating seedlings represents a major challenge to plant breeding for many crops, including cucurbits. Due to the lack of effective chemicals for virus disease control, many researchers have focused on understanding the interaction mechanisms between viruses and hosts in order to develop potential resistant materials or breed resistant cultivars [1][2]. As a seed-borne virus, cucumber green mottle mosaic virus (CGMMV), which is a member of the Tobamovirus genus, has spread worldwide via the international seed trade. CGMMV has a 6.4 kb single-stranded, positive-sense RNA genome containing four open reading frames (ORFs) [3]. Two co-terminal ORFs (ORFs 1 and 2) encode two proteins, a predominant 129 kDa protein and a 186 kDa readthrough protein, responsible for RNA replication. ORF 3 encodes a 29 kDa protein involved in viral cell-to-cell movement (movement protein (MP)), and ORF 4 encodes a 17.4 kDa coat protein (CP) [4] required for viral packaging and transmission. CGMMV primarily infects Cucurbitaceae members, causing mottling, mosaic patterns, and brown necrotic lesions on the stems, leaves, and fruits. Symptoms vary between different cucurbit crop species and cultivars of the same species [5]. Generally, the transmission rate of CGMMV in contaminated watermelon seeds is 1–10%, and transmission is greater than 12% in cucumber seeds [6][7]. Similar to many Tobamoviruses, CGMMV virions are stable, and virus particles on surfaces remain infectious for more than a year and can spread via mechanical transmission without insect vectors. When CGMMV-contaminated seeds are sown, virions from the seed coat can infect the germinating seedlings through tiny wounds that form during early growth, causing yield losses as high as 15% and >50% in cucumber and watermelon, respectively [8][9]. Therefore, effective methods to control CGMMV disease are prerequisites for cucurbits production. Currently, the most common method for virus control is through chemical and biological approaches. Sodium hypochlorite is the most effective disinfectant/virucidal chemical against Tobamoviruses. However, although disinfectants can remove CGMMV from the outer seed coat, they cannot remove virions present inside the seed [7]. Breeding cucurbits that are genetically resistant to CGMMV is hindered by the scarcity of resistance genes and instability due to temperature sensitivity [10]. Although cucumber plants can be protected from highly infectious strains via cross-protection, diseases can still emerge due to interactions with other viruses, mixed infections, or recombination [11], and effective biological measures are still needed for disease prevention and control.
RNA interference (RNAi) represents a practical approach for developing plant resistance and involves the silencing of genes via sequence-specific suppression or cleavage of complementary mRNA by small RNAs (sRNAs) [12][13]. Currently, virus-induced gene silencing (VIGS) and hairpin RNA-based silencing represent the main antiviral RNAi approaches used [14]. Despite their effectiveness, their antiviral effect is still affected by a large number of derived small interfering RNAs (siRNAs), which leads to off-target effects [15]. To reduce the occurrence of such events, second-generation RNAi strategies based on artificial sRNAs, such as artificial miRNA (amiRNA) and synthetic trans-acting siRNA (syn-tasiRNA)-mediated gene silencing, has been used to induce resistance to viral infection and modify several crops to obtain ideal agronomic traits [16][17][18]. Artificial sRNAs are produced in plants by expressing a functional miRNA or tasiRNA precursor containing modified miRNA/miRNA* or tasiRNA sequences, respectively. By using an overlapping PCR approach, the amiRNA precursor, which is obtained by replacing the original miRNA/miRNA* duplex region, is processed by Dicer-Like 1 (DCL1) to produce amiRNA/amiRNA* duplexes. The amiRNA strand is recruited by ARGONAUTE (AGO) proteins to form miRNA-induced silencing complexes to mediate post-transcription gene silencing or translation repression (Figure 1A) [19]. syn-tasiRNA precursors are first cleaved by a miRNA–AGO complex. Then, there is the conversion of one of the cleavage products into double-stranded RNA (dsRNA) by RNADEPENDENT RNA POLYMERASE 6 and sequential processing by DCL4 of the dsRNA into 21-nucleotide phased syn-tasiRNAs registered with the miRNA-guided cleavage site (Figure 1B) [20]. amiRNAs only recognize target sequences containing less than five mismatches, which confers high silencing specificity [21][22]. Multiple endogenous plant genes sharing a short conserved sequence can be silenced simultaneously. In addition, amiRNA-mediated viral resistance remains effective even at low temperatures [23]. syn-tasiRNAs can co-express a single precursor of several syn-tasiRNAs targeting multiple sites in a single viral RNA or different viral RNAs, inducing more effective, durable, and broad antiviral resistance. However, viruses can inhibit antiviral RNA silencing by expressing silencing suppressors, which support infection and reproduction; the mode of action of the silencing suppressor varies between different viruses [24]. The CMV-2b protein, which was the first identified silencing suppressor, can bind to double-stranded sRNA in vivo and in vitro [25][26]. The coat protein P38 in turnip crinkle viruses has been suggested to inhibit DCL4-mediated siRNA processing [27]. By comparing the SH and SH33b strains of CGMMV, it was found that a single amino acid substitution from E to G at aa position 480 in the intervening region of the 129 K protein is responsible for the impaired RNA-silencing activity (SSA) and siRNA-binding capability (SBC) of SH33b, resulting in attenuated symptoms [28]. However, it is unknown whether the accumulation of CGMMV caused by seed-based transmission in cucumber plants may mediate binding to amiRNA or affect the expression of amiRNA and, thereby, affect the antiviral effect of amiRNA. Therefore, it is vital to investigate whether an amiRNA-based system can induce resistance to CGMMV infections in cucumber plants in this study.
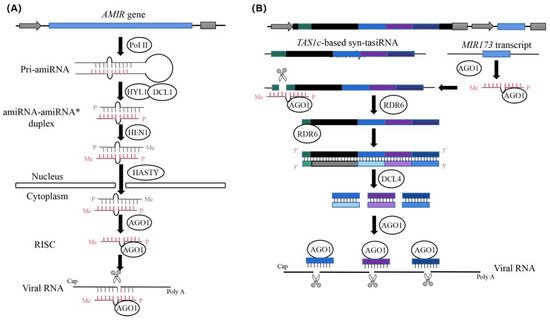
Figure 1. The diagram of the process of antivirus artificial microRNA (amiRNA) and synthetic trans-acting small interfering RNA (syn-tasiRNA) production (revised from Jones-Rhoades et al. and Carbonell et al.). (A) Diagram of the biogenesis and activities of artificial microRNA (amiRNA) products. Pri-amiRNAs are transcribed by RNA polymerase II(Pol II). The amiRNA-amiRNA* duplex produced by DCL1 is then exported to the cytoplasm, possibly through the action of the plant exportin 5 ortholog HASTY. The guide amiRNA strand is then incorporated into AGO proteins to carry out the silencing reactions. (B) Diagram of the biogenesis and activities of synthetic trans-acting small interfering RNA (syn-tasiRNA) products. Three different antivirus syn-tasiRNAs (in light or dark blue or purple boxes) were inserted into the TAS1c gene from Arabidopsis thaliana (in black). The miR173 target site (TS) is shown with a green square box. A cassette, including the A. thaliana MIR173 precursor (in blue) to generate miR173, was inserted downstream of the syn-tasiRNA cassette. Specific cleavage sites in target sites located in viral RNAs are indicated with black arrows, with TS coordinates indicated in brackets.
2. Design of amiRNAs against Multiple CGMMV Strains
To confer resistance against different strains of CGMMV, amiRNAs were designed to target conserved regions of 25 CGMMV strains based on data obtained from the National Center for Biotechnology Information (NCBI). The genome was screened using a 21-nucleotide window (TNNNNNNNNNNNNNNNNNNNN), and the published amiRNA selection criteria from the WMD3 output list (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi, accessed on 20 March 2019) were applied, including up to two mismatches at position 1 or 15–21, absolute hybridization energy between −35 and −38 kcal/mole, and with a dGamiR-target/dGperfect-match value >80% [29]. To reduce the unintended effects, potential amiRNAs targeting the identified target sites were investigated for potential off-target activities via WMD3 target search (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi/page=TargetSearch, accessed on 21 March 2019) and cucumber (Chinese long) genome v2 database (http://www.cucurbitgenomics.org, accessed on 21 March 2019) analysis. Based on these standards, we selected six amiRNAs targeting different positions, of which three targeted the Rep genes, two targeted MPs, and one targeted the CP, which were designated amiR1-Rep, amiR2-Rep, amiR3-Rep, amiR4-MP, amiR5-MP, and amiR6-CP; amiR-GUS (GUS, β-glucuronidase) was selected as a control (Figure 2). We found no cucumber endogenous genes that functioned as potential targets when two mismatches were allowed.
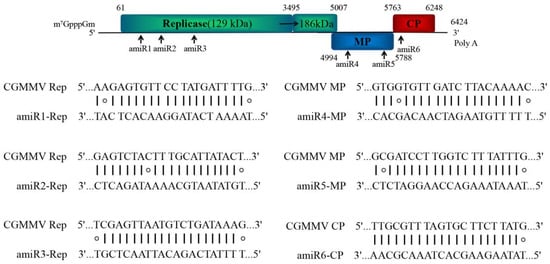
Figure 2. Schematic diagram of amiRNA designed for silencing the CGMMV genes coding for replicase proteins, movement protein (MP), and coat protein (CP). amiRNA, artificial microRNA; CGMMV, cucumber green mottle mosaic virus.
Similar to our previous study, overlapping polymerase chain reaction (PCR) techniques were applied to generate amiRNA precursors by replacing the original miRNA/miRNA* duplex in ath-miR156, ath-miR164, and ath-miR171 backbones [30]. By using the mFold program, the computational prediction indicated that the RNA secondary structure of all amiRNA precursor constructs possessed correct folding parameters.
3. Expression and Anti-CGMMV Activity of amiRNAs in Nicotiana Benthamiana
In previous studies, we found that amiRNA expression and antiviral activity were correlated [30]. Therefore, we aimed to check whether the expression levels of the anti-CGMMV amiRNA correlated with the antiviral activity using transient expression assays in N. benthamiana. Here, we agroinfiltrated each construct into the plants to compare the expression of each amiRNA; –amiR-GUS was used as a control. Northern blot analysis of RNA preparations obtained three days post-agroinfiltration revealed that all amiRNAs were expressed. Among them, amiR4-MP had the highest expression level, while the expression level of amiR1-Rep and amiR5-MP were similar with the control; quantitative reverse transcription-PCR (qRT-PCR) results also confirmed that the expression levels of amiR4-MP and amiR6-CP were approximately 20- and 10-fold higher than those of the control, respectively (Figure 3A). The trend of the amiRNA expression level in CGMMV-infected N. benthamiana plants at 3, 10, and 15 days post-inoculation (dpi) was analyzed using a previously described assay [30]. In general, the expression level of amiRNA showed a downward trend, and the decrease in expression level slowed down after 10 dpi and was maintained at a certain level (Figure 3B).
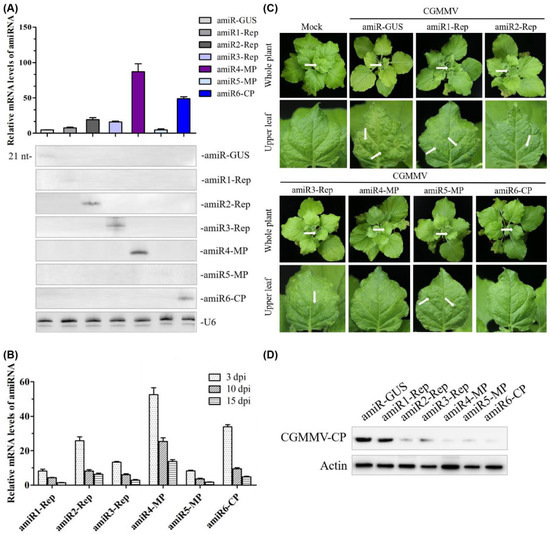
Figure 3. Functional analysis of the anti-CGMMV amiRNA in Nicotiana benthamiana. (A) amiRNA expression level detected by qRT-PCR (upper panel) and Northern blot hybridization (lower panel); from left to right in upper and lower panels: amiR-GUS, amiR1-Rep, amiR2-Rep, amiR3-Rep, amiR4-MP, amiR5-MP, and amiR6-CP. RNA isolated from agroinfiltrated leaves at 3 dpi. Each biological replicate represents a pool of two agroinfiltrated leaves from the same plant. U6 hybridization serves as a Northern blot loading control. (B) Relative expression of amiR1–amiR6 at 3, 10, and 15 dpi in CGMMV-infected N. benthamiana plants. Error bars represent the mean of three biological replicates ± standard deviation (SD). (C) Photographs of CGMMV-infected whole plants and upper leaves at 20 dpi. Characteristic symptoms of CGMMV-induced mild mottle and mosaic are indicated with white arrows shown in upper leaves, as shown below. White arrows on whole plants indicate the upper leaves. (D) Western blot hybridization detection of CGMMV coat protein (17.4 kDa) accumulation in infected upper leaves at 20 dpi. dpi, days post-inoculation. amiR1-Rep, amiR2-Rep, and amiR3-Rep mean the amiRNA targeted three different regions of the Rep gene of CGMMV, respectively; amiR4-MP and amiR5-MP mean the amiRNA targeted two different regions of the MP gene of CGMMV, respectively; amiR6-CP means the amiRNA targeted the CP gene of CGMMV; amiR-GUS means the amiRNA targeted the GUS gene.
To compare the antiviral activity of different amiRNAs, six agroinfiltrated leaves from independent plants were inoculated with CGMMV after 3 days of amiRNA treatment. We monitored the appearance of characteristic CGMMV-induced symptoms in the inoculated tissues (necrotic lesions) and upper non-inoculated tissues (deformed leaves and mottle) and detected the protein accumulation of CGMMV in upper non-inoculated leaves at 20 dpi by Western blot. It was shown that the upper non-inoculated tissues in all plants infiltrated with amiR-GUS displayed strong leaf green mottle. Plants agroinfiltrated with amiR4-MP and amiR6-CP did not show visible symptoms (Figure 3C) and displayed reduced levels of CGMMV accumulation (Figure 3D). However, plants infiltrated with amiR1-Rep and amiR5-MP showed multiple green mottles on the upper leaves at 20 dpi. The accumulation of viruses in the upper leaves of the amiR1-Rep-infiltrated plants was significantly higher than that in amiR5-MP-infiltrated lines (Figure 3C,D).
4. Identify the Most Effective amiRNA for Generating Polycistronic Constructs
To confirm the antiviral activity of each amiRNA more rapidly and directly, we generated a transient in vivo sensor system based on a previous study [23]. By introducing the target sites of amiRNA into the 3′ end of the green fluorescent protein (GFP) reporter gene, we prepared sensor constructs, designated pGFPamiR1/4, pGFPamiR2/5, and pGFPamiR3/6, containing the 21-nucleotide target sequences in the CGMMV genome for amiR1-Rep–amiR6-CP, respectively (Figure 4A). Agrobacterium-mediated co-transformation experiments were performed by co-infiltrating the GFP sensor constructs with the particular amiRNAs.
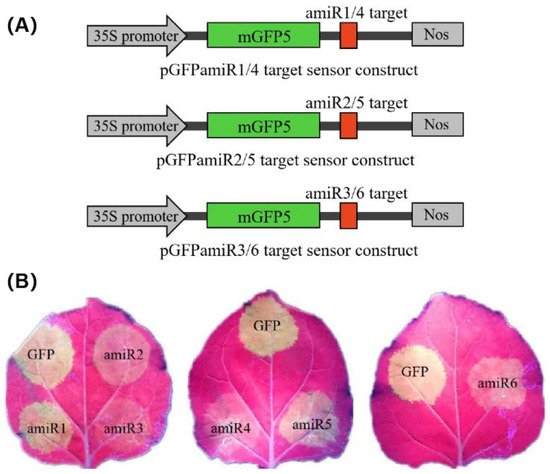
Figure 4. amiRNA functional assays for detecting transient co-expression of amiRNAs and amiR-GFP sensors. (A) Schematic representation of three pGFPamiR target sensor constructs, each containing two (amiR1 and amiR4, amiR2 and amiR5, amiR3 and amiR6) amiRNA target sites in the 3′ untranslated region of the mGFP5 reporter gene, under the control of the cauliflower mosaic virus 35S promoter. (B) Biological activity of amiRNAs in transient co-infiltration assays. The pGFPamiR target sensor constructs were co-infiltrated with the corresponding amiRNA constructs into Nicotiana benthamiana leaves. As a control, the original pEG100-miR171 and either of the pGFPamiR sensor constructs were co-infiltrated. The reduced GFP fluorescence indicates target cleavage by the biological activity of the individual amiRNAs, whereas bright fluorescence indicates the disabled function of the amiRNAs. The three biological repeats of the transient assays showed similar results. GFP, green fluorescent protein.
Based on the fluorescence intensity, it was found that amiR2-Rep, amiR3-Rep, amiR4-MP, and amiR6-CP efficiently inhibited the activity of their target regions. Among them, amiR3-Rep, which was expressed at a 3-fold higher level than those of the control in N. benthamiana at 3 dpa (Figure 3A), almost completely inhibited the expression of the GFP sensor construct. In contrast, amiR1-Rep and amiR5-MP, which showed a similar expression level to the control in N. benthamiana did not significantly inhibit the expression of the GFP sensor construct (Figure 4B). To evaluate the specificity of each amiRNA to its target, we set up a mismatch experiment group containing the anti-CGMMV amiRNA and GFP sensor constructs. By co-infiltrating the amiR1-Rep with pGFPamiR2/5, amiR2-Rep with pGFPamiR3/6, amiR3-Rep with pGFPamiR1/4, amiR4-MP with pGFPamiR2/5, amiR5-MP with pGFPamiR3/6, and amiR6-CP with pGFPamiR1/4, respectively, it was found that there is no difference in the fluorescence intensity between the mismatched group and the blank control. The potential biological activity of the amiRNA candidates suggested that their ability to silence the virus can be identified using GFP sensors and the amiRNA-mediated resistance effect detected in N. benthamiana to comprehensively compare the effect of amiRNA and not only judged according to the expression level of amiRNA.
Based on the performance of amiRNA in silencing the virus, we selected amiR2-Rep, amiR4-MP, and amiR6-CP to generate polycistronic constructs, which generated multiple amiRNAs from a single transcript and were inspired by the polycistronic miRNAs found in nature [31]. The mFold program was used to predict the secondary RNA structure of the resulting amiRNA constructs, revealing that the individual amiRNA backbones could fold correctly (Figure 5A). To test the biological activity of the polycistronic amiRNA construct, we infiltrated it into N. benthamiana and inoculated it with CGMMV. The expression level of amiRNAs in the polycistronic constructs was lower than that of amiRNAs alone at the same period (Figure 5B). However, there were no symptoms of CGMMV in the upper leaves at 20 dpi (Figure 5C).
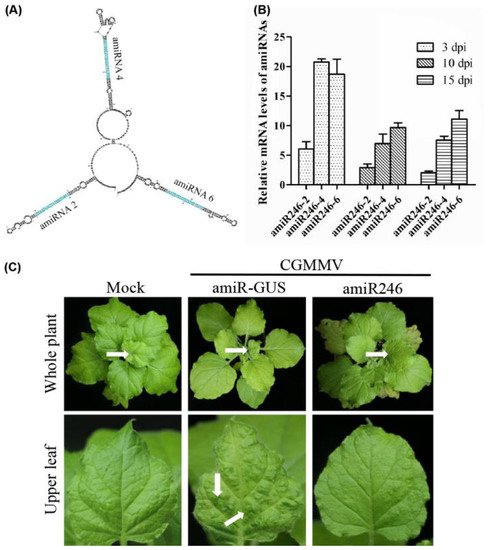
Figure 5. Biological activity of the polycistronic amiR246 to CGMMV in Nicotiana benthamiana transient assays. (A) Predicted secondary structure of polycistronic amiRNA (amiR246) via mFold analysis. (B) Expression levels of three amiRNAs in the polycistronic structure at 3, 10, and 15 dpi after inoculating with CGMMV in N. benthamiana plants. For each amiRNA, three biological replicates were evaluated using quantitative reverse transcription-polymerase chain reaction, and three technical replicates were performed for each sample. Error bars represent the mean of three biological replicates ± standard deviation (SD). (C) Photographs of the upper leaves and whole plants were taken at 20 dpi. Characteristic symptoms of CGMMV-induced mild leaf mottle and mosaic are indicated with white arrows. White arrows on whole plants indicate the upper leaves.
This entry is adapted from the peer-reviewed paper 10.3390/ijms222212237
References
- Wang, D.W.; Maule, A.J. A Model for Seed Transmission of a Plant Virus: Genetic and Structural Analyses of Pea Embryo Invasion by Pea Seed-Borne Mosaic Virus. Plant Cell 1994, 6, 777–787.
- Aranda, M.A.; Escaler, M.; Wang, D.; Maule, A.J. Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc. Natl. Acad. Sci. USA 1996, 93, 15289–15293.
- Ugaki, M.; Tomiyama, M.; Kakutani, T.; Hidaka, S.; Kiguchi, T.; Nagata, R.; Sato, T.; Motoyoshi, F.; Nishiguchi, M. The complete nucleotide sequence of cucumber green mottle mosaic virus (SH strain) genomic RNA. J. Gen. Virol. 1991, 72, 1487–1495.
- Liu, Y.; Wang, Y.A.; Wang, X.F.; Zhou, G.H. Molecular Characterization and Distribution of Cucumber green mottle mosaic virus in China. J. Phytopathol. 2009, 157, 393–399.
- Dombrovsky, A.; Tran-Nguyen, L.T.; Jones, R.A. Cucumber green mottle mosaic virus: Rapidly Increasing Global Distribution, Etiology, Epidemiology, and Management. Annu. Rev. Phytopathol. 2017, 55, 231–256.
- Liu, H.W.; Luo, L.X.; Li, J.Q.; Liu, P.F.; Chen, X.Y.; Hao, J.J. Pollen and seed transmission of Cucumber green mottle mosaic virus in cucumber. Plant Pathol. 2014, 63, 72–77.
- Reingold, V.; Lachman, O.; Blaosov, E.; Dombrovsky, A. Seed disinfection treatments do not sufficiently eliminate the infectivity of Cucumber green mottle mosaic virus (CGMMV) on cucurbit seeds. Plant Pathol. 2015, 64, 245–255.
- Fletcher, J.T.; George, A.J.; Green, D.E. Cucumber Green Mottle Mosaic Virus, its Effect on Yield and its Control in the Lea Valley, England. Plant Pathol. 1969, 18, 16–22.
- Reingold, V.; Lachman, O.; Koren, A.; Dombrovsky, A. First report of Cucumber green mottle mosaic virus (CGMMV) symptoms in watermelon used for the discrimination of non-marketable fruits in Israeli commercial fields. New Dis. Rep. 2013, 28, 11.
- Sugiyama, M.; Ohara, T.; Sakata, Y. A New Source of Resistance to Cucumber Green Mottle Mosaic Virus in Melon. J. Jpn. Soc. Hortic. Sci. 2006, 75, 469–475.
- Crespo, O.; Robles, C.; Ruiz, L.; Janssen, D. Antagonism of Cucumber green mottle mosaic virus against Tomato leaf curl New Delhi virus in zucchini and cucumber. Ann. Appl. Biol. 2020, 176, 147–157.
- Baulcombe, D.C. RNA silencing in plants. Nat. Cell Biol. 2004, 431, 356–363.
- Cillo, F.; Palukaitis, P. Transgenic resistance. Adv. Virus Res. 2014, 90, 35–146.
- Burch-Smith, T.; Anderson, J.C.; Martin, G.B.; Dinesh-Kumar, S.P. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004, 39, 734–746.
- Xu, P.; Zhang, Y.J.; Kang, L.; Roossinck, M.J.; Mysore, K.S. Computational estimation and experimental verification of off-target silencing during post transcriptional gene silencing in plants. Plant Physiol. 2006, 142, 429–440.
- Niu, Q.-W.; Lin, S.-S.; Reyes, J.L.; Chen, K.-C.; Wu, H.-W.; Yeh, S.-D.; Chua, N.-H. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 2006, 24, 1420–1428.
- Butardo, V.; Fitzgerald, M.; Bird, A.; Gidley, M.J.; Flanagan, B.M.; Larroque, O.; Resurreccion, A.P.; Laidlaw, H.K.C.; Jobling, S.; Morell, M.K.; et al. Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA- and hairpin RNA-mediated RNA silencing. J. Exp. Bot. 2011, 62, 4927–4941.
- Chi, M.; Bhagwat, B.; Lane, W.D.; Tang, G.; Su, Y.; Sun, R.; Oomah, B.D.; Wiersma, P.A.; Xiang, Y. Reduced polyphenol oxidase gene expression and enzymatic browning in potato (Solanum tuberosum L.) with artificial microRNAs. BMC Plant Biol. 2014, 14, 62.
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53.
- Carbonell, A.; Lisón, P.; Daròs, J. Multi-targeting of viral RNAs with synthetic trans -acting small interfering RNAs enhances plant antiviral resistance. Plant J. 2019, 100, 720–737.
- Schwab, R.; Ossowski, S.; Riester, M.; Warthmann, N.; Weigel, D. Highly Specific Gene Silencing by Artificial MicroRNAs inArabidopsis. Plant Cell 2006, 18, 1121–1133.
- Khraiwesh, B.; Ossowski, S.; Weigel, D.; Reski, R.; Frank, W. Specific Gene Silencing by Artificial MicroRNAs in Physcomitrella patens: An Alternative to Targeted Gene Knockouts. Plant Physiol. 2008, 148, 684–693.
- Kis, A.; Tholt, G.; Ivanics, M.; Varallyay, E.; Jenes, B.; Havelda, Z. Polycistronic artificial miRNA-mediated resistance to Wheat dwarf virus in barley is highly efficient at low temperature. Mol. Plant Pathol. 2016, 17, 427–437.
- Vargason, J.M.; Szittya, G.; Burgyán, J.; Hall, T.M. Size Selective Recognition of siRNA by an RNA Silencing Suppressor. Cell 2003, 115, 799–811.
- González, I.; Rakitina, D.; Semashko, M.; Taliansky, M.; Praveen, S.; Palukaitis, P.; Carr, J.; Kalinina, N.; Canto, T.; Semashko, M. RNA binding is more critical to the suppression of silencing function of Cucumber mosaic virus 2b protein than nuclear localization. RNA 2012, 18, 771–782.
- Hamera, S.; Song, X.; Su, L.; Chen, X.; Fang, R. Cucumber mosaic virus suppressor 2b binds to AGO4-related small RNAs and impairs AGO4 activities. Plant J. 2012, 69, 104–115.
- Deleris, A.; Gallego-Bartolome, J.; Bao, J.; Kasschau, K.D.; Carrington, J.C.; Voinnet, O. Hierarchical Action and Inhibition of Plant Dicer-Like Proteins in Antiviral Defense. Science 2006, 313, 68–71.
- Chen, H.; Ino, M.; Shimono, M.; Wagh, S.G.; Kobayashi, K.; Yaeno, T.; Yamaoka, N.; Bai, G.; Nishiguchi, M. A Single Amino Acid Substitution in the Intervening Region of 129K Protein of Cucumber Green Mottle Mosaic Virus Resulted in Attenuated Symptoms. Phytopathology 2020, 110, 146–152.
- Li, J.-F.; Chung, H.S.; Niu, Y.; Bush, J.; McCormack, M.; Sheen, J. Comprehensive Protein-Based Artificial MicroRNA Screens for Effective Gene Silencing in Plants. Plant Cell 2013, 25, 1507–1522.
- Liang, C.Q.; Hao, J.J.; Li, J.Q.; Baker, B.; Luo, L.X. Artificial microRNA-mediated resistance to cucumber green mottle mosaic virus in Nicotiana benthamiana. Planta 2019, 250, 1591–1601.
- Merchan, F.; Boualem, A.; Crespi, M.; Frugier, F. Plant polycistronic precursors containing non-homologous microRNAs target transcripts encoding functionally related proteins. Genome Biol. 2009, 10, R136.
This entry is offline, you can click here to edit this entry!
