Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Physiology
Vitamins are essential compounds obtained through diet that are necessary for normal development and function in an organism. One of the most important vitamins for human physiology is vitamin A, a group of retinoid compounds and carotenoids, which generally function as a mediator for cell growth, differentiation, immunity, and embryonic development, as well as serving as a key component in the phototransduction cycle in the vertebrate retina.
- vitamin A transporters
- all-trans retinol
- retinyl esters
- LRAT
- STRA6
- RBPR2
- RBP4
1. Mechanisms Involving Intestinal Absorption of Provitamin A Carotenoids and Preformed Vitamin A
Before the important roles of vitamin A transporters can be discussed, the general schematic of macroscale vitamin A interconversions and movements within the cell and circulation must first be established. The base isoprenoid structure of vitamin A in all forms found in biological processes are derived from two dietary sources: β-carotene from plant sources and retinyl palmitate from animal sources [1][2][3]. Besides the method of intake for these two vitamin A sources, the subsequent pathways after absorption converge. For β-carotene, this macromolecule enters intestine epithelial cells through the scavenger receptor class B, type 1 (SCAR-B1/SR-B1) transporter, where intracellular β-carotene is then cleaved into two molecules of all-trans retinal by β-carotene monooxygenase 1 (BCO1/BCMO1), which is then reduced into all-trans retinol in the cytosol [4]. For extracellular retinyl palmitate, the molecule is first hydrolyzed by extracellular retinyl esterases into all-trans retinol, the alcohol then diffuses into the epithelial cell. The all-trans retinol from both sources binds to cellular retinol binding protein 2 (CRBP2). The retinol-CRBP2 complex then interacts with lecithin retinol acyltransferase (LRAT) and converts the retinol into retinyl esters and dissociates with CRBP2 [4][5][6]. These retinyl esters are then packaged into nascent chylomicrons along with other lipids and excreted into general circulation through exocytosis. It should be mentioned that most retinyl esters stay with the nascent chylomicron throughout its process in converting into a chylomicron remnant [7]. Around 70% of these chylomicrons are absorbed into the liver for storage. The remaining chylomicron remnants are absorbed into peripheral organs, thus acting as another pathway for vitamin A delivery in extrahepatic tissues [7][8].
The tissue storage of vitamin A is facilitated by hepatocyte-associated hepatic stellate cells. These two cell types function together and can respond to stimuli indicating low vitamin A in body systems; as such, the hepatocyte-associated hepatic stellate cells are capable of both in-taking vitamin A for storage and releasing vitamin A into lymphatic capillaries and blood vessels. It should be mentioned that the mechanism of function for detection of low concentrations of vitamin A in the peripheral organs is unknown and individual peripheral organs such as the eye are hypothesized to be able to independently signal hepatocytes for vitamin A release [1][2][3]. The arrived retinyl esters are first hydrolyzed by intracellular retinyl ester hydrolases into cellular retinol-binding protein 1 (CRBP1) bound all-trans retinol, which then enters the hepatic stellate cell and transformed into storage capable retinyl esters by LRAT. Excretion of vitamin A first involves hydrolysis of storage retinyl esters into all-trans retinol, which is then excreted through the hypothesized efflux capabilities of the retinol-binding protein 4 receptor 2 (RBPR2) in the intestines. Ejected all-trans retinol then associates with RBP4 and transthyretin to form a complex that allows movement through lymphatic and cardiovascular vessels [1][2][3] (Figure 1).
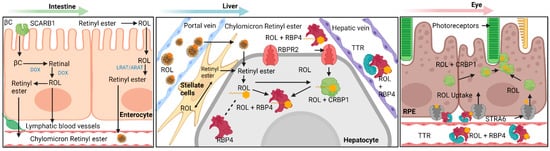
Figure 1. Overview diagram detailing the various pathways within vitamin A transport and metabolism into biologically useful forms. Generally, this process can be separated into four different categories: absorption, storage, release, and uptake. A succinct summary of these four categories is shown here for vitamin A: absorption of provitamin A into intestinal epithelial cells and initial processing into chylomicron bound retinyl esters, storage of retinyl esters within hepatic stellate cells, release of storage retinyl esters as RBP4 bound retinol into the bloodstream and uptake by the retinal pigment epithelium. Note the importance of STRA6 and RBPR2 as major facilitators in the transport of RBP4 bound retinol. βC—β-Carotene; SCARB1—Scavenger Receptor Class B, Type 1; LRAT—Lecithin Retinol Acyltransferase; ARAT—Acyl-CoA Retinol Acyltransferase (ARAT); ROL—All-Trans Retinol; STRA6 – Stimulated by Retinoic Acid 6; RBPR2—Retinol Binding Protein 4 Receptor 2; RBP4—Retinol Binding Protein 4; TTR—Transthyretin; CRBP1—Cellular Retinol Binding Protein 1; CRBP2—Cellular Retinol Binding Protein 2; RPE—Retinal Pigment Epithelium. Created with BioRender.com.
2. Uptake of Carotenoids—SR-B1
SCARB1 or SR-B1, is a 509 amino acid integral membrane protein that facilitates the uptake of many different macromolecules into epithelial cells. Through nuclear magnetic resonance microscopy (NMR), it was found that a leucine zipper dimerization motif found in the trans-membrane domain C-terminal was integral to its ability to bind lipoproteins [9]. As such, SCARB1 is an important regulator of cholesterol metabolism and lipid metabolism, functioning as a receptor for low density, very low density, and high-density lipoproteins [10]. Additionally, SCARB1 can also serve as a transporter for vitamins, including tocopherols, and carotenoids such as β-carotene and xanthophylls [10][11].
The importance of SCARB1 in carotenoid transport was demonstrated through the seminal work from the von Lintig Lab. Fruit flies containing a nonsense mutation in neither inactivation nor afterpotential D (ninaD) gene eliminates the expression of the fruit fly SCARB1 analog. These mutant flies displayed significantly lower carotenoid composition in the carotenoid heavy areas of the trunk and head, as well the presence of immature rhodopsin in the retina. Furthermore, a diet supplemented with preformed vitamin A or significantly high amounts of β-carotene was shown to be able to allow for rhodopsin maturation in ninaD flies, with both diets bypassing the lack of functional SCARB1 [12].
2.1. Carotenoid Cleaving Enzymes—BCO1, BCO2
BCO1 and BCO2 belongs to an enzyme family called carotenoid cleavage oxygenases (CCOs). CCOs are characterized by their ability to cleave the carotenoid polyene backbone with high stereoselectivity and regioselectivity, thus cleaving only selected polyenes at specific sites leaving specific products with very high fidelity [13][14][15]. Due to the hydrophobic nature of its substrates and its storage within hydrophobic liposomes, CCOs contain external regions of α-helices with hydrophobic residues that allow for its interaction with phospholipid bilayers and carotenoid substrates. Another structural characteristic of note is the presence of hydrophobic “tunnels” that allow for the entrance of the hydrophobic carotenoid into the catalytic core [16].
All enzymes in the CCO enzyme family contain a Fe2+ in the catalytic center fixed by four highly conserved His residues, which are in turn fixed by three highly conserved Glu residues. The iron catalytic core then activates oxygen for one of two theorized mechanisms of polyene cleavage. The monooxygenase reaction hypothesizes a two-step reaction through an epoxide intermediate and a trans-diol intermediate, while the dioxygenase reaction involves a one-step reaction through a highly unstable dioxetane intermediate [16].
In the β-carotene metabolic pathway, BCO1 catalyzes the cleavage of β-carotene taken in through SCARB1 into two molecules of retinal through symmetrical cleavage of β-carotene. This retinal will then serve as the precursor for all carotenoids found in biological processes, such as retinyl esters for storage in the liver, all-trans retinoic acid for use as a ligand for RAR-RXR transcription factors, or 11-cis retinol for entry into the visual cycle [13].
While BCO1 functions to cleave β-carotene symmetrically for conversion into biologically useful forms, the information known for BCO2 is comparatively much less. Catalytically, BCO2 is different from BCO1 in that it cleaves β-carotene asymmetrically, creating what is known as apocarotenals. BCO2 also seems to preferentially bind xanthophylls over carotenes. These apocarotenals serve a multitude of functions that are just now being elucidated by various laboratories. Some of these functions include regulation in mitochondrial apoptosis and working in conjunction with BCO1 to generate retinoids from asymmetric β-cryptoxanthin [17].
2.2. An Intestinal Transcription Factor Regulates Vitamin A Absorption—ISX
The pathway for absorption of carotenoids is negatively regulated by the homeodomain transcription factor (ISX). The expression of ISX is directly stimulated by the retinoic acid receptor (RAR) and the downstream retinoic acid metabolite, which leads to the downregulation of SCARB1 and BCO1. The repression of these two fundamental proteins in the retinoid pathway by retinoic acid-induced ISX constitutes a negative feedback loop that reduces provitamin A intake while retinoid is plentiful [18].
3. Transport of All-Trans Retinol to Extrahepatic Organs—RBP4 and Retinol
All-trans retinol is the fundamental transport form of vitamin A; additionally, all functional retinoids and vitamin A metabolites are derived from retinol. Alongside retinyl esters, retinol is the most abundant form of vitamin A. Retinyl esters serves as the main storage form of retinol, acts as the primary transport form of newly arrived vitamin A, and acts as the precursor for vitamin A metabolites [19]. The transport of hepatic retinol within the serum is facilitated through its binding to retinol-binding protein 4 (RBP4). RBP4, the transport protein responsible for hepatic retinol transport, is primarily expressed in hepatic liver tissue, where it forms a holo-enzyme complex with the retinol substrate and transthyretin (TTR) that gets mobilized out of the hepatocyte and into circulation. The majority (about 85%) of circulating RBP4 is in a holoenzyme complex with retinol, with the remaining 15% in an apo-RBP4 state [20]. Adipose tissue can express RBP4, but they are not a major source of the holo-enzyme circulating in the body, such as those produced in liver hepatocytes. Mice that have liver RBP4 knocked out displayed a significant decrease in serum RBP4 while maintaining normal adipose production of the transport protein [21]. The complex formed between holo-RBP4 and TTR is shown to enhance the secretion of RBP4 from the liver into circulation, as well as to help stabilize the complex and reduce its likelihood of renal filtration out of the body [22].
The concentration of RBP4/Retinol lies in a very narrow and regulated range of around 2–3 uM in humans and 1 uM in mice [23], where fluctuations from this concentration range can lead to dysfunction and disorder. Elevated levels of holo-RBP4 in circulation are correlated to an increase in insulin tolerance, subsequent type 2 diabetes, and obesity, as well as the development of nonalcoholic fatty liver disease (NAFLD), among other metabolic diseases based on several case studies involving observational studies of human patients with RBP4 and retinol deficiencies in circulation, and molecular studies using mouse and cell culture models [20]. However, these findings are controversial, with some counter studies suggesting that there is no link between increased circulating RBP4 and insulin resistance, type 2 diabetes, and NAFLD [20][24]. Decreased concentrations of circulating RBP4 have been linked to night-blindness, which was noted in a couple of case studies where individuals experienced impaired vision and retinal dystrophy from low RBP4 serum concentration, which was likely due to mutations found in RBP4 or the complete lack of the protein. Furthermore, reduced RBP4 serum concentration does not appear to lead to any abnormal phenotype other than impaired visual function [24].
4. Known Vitamin A Transporters/RBP4 Receptors
4.1. Stimulated by Retinoic Acid 6 (STRA6)
Transport of retinoids in and out of cells has been hypothesized to be facilitated by cell surface receptor transport proteins since the initial report of a cell surface receptor for RBP in the 1970s [25]. Despite their initial reports in the 1970s, the cell surface receptor for RBP bound retinol was not characterized for another three decades until the Sun lab in UCLA identified stimulated by retinoic acid 6 (STRA6) in the retinal pigment epithelium (RPE) as a major cell surface transporter for holo-RBP/Retinol in 2007 [26]. The gene for STRA6 was originally identified in 1995 with an unknown function [27]. STRA6 was later postulated to be an integral membrane protein and was found to be highly expressed in rodent embryos and, in rodent adults, the eye (RPE), brain, kidneys, spleen, testes, and female genital tract [28].
The proper function and characterization of STRA6 as the major RBP-bound retinoid transport protein was elucidated a decade later with the work of Kawaguchi et al. in 2007, where they found the membrane protein to regulate the uptake of RBP-bound retinol in bovine RPE cells (Figure 1). STRA6 binds to the holo-RBP complex with high affinity (Kd = 59 nM) and shuttles the RBP-bound retinol across the membrane [26]. STRA6 transports retinol bidirectionally depending on RBP’s and LRAT concentrations intracellularly and holo/apo-RBP’s extracellularly, with influx occurring in the presence of LRAT, apo-CRBP1, and holo-RBP, and efflux in the presence of holo-CRBP1 and apo-RBP [29]. Much of these previous findings were made without knowledge of the structure of STRA6, which was believed to be novel for an integral protein until 2016.
Mutations in STRA6 can lead to a myriad of diseases and phenotypes. STRA6 mutations during development can lead to anophthalmia, microphthalmia, and other symptoms that overlap with phenotypes associated with Matthew-Wood syndrome [30]. Matthew-Wood syndrome is a rare congenital disease associated with microphthalmia, mental deficiencies, and various organ deformities [31]. However, these phenotypes can range from mild to severe depending on the mutations of STRA6 and are not explicitly caused by STRA6 mutations but by an excess of retinoic acids in some cases [31]. The associations between mutations in STRA6 and incidence of Matthew-Wood syndrome have been used to create animal models for Matthew-Wood Syndrome in later research, such as with the work of the von Lintig lab in 2008, where they found that excess holo-RBP4 in circulation disrupts vitamin A uptake and causes developmental abnormalities, such as those seen in Matthew-Wood Syndrome, using a morpholino approach to generate a Stra6-deficient zebrafish model [32]. Further research on STRA6 mutations was conducted using mammalian models with STRA6 knockout mice. In 2012, the Bok lab in UCLA generated a STRA6 knockout mouse model to find changes in their visual function and development, finding that the mutant mice had phenotypic differences from controls such as reduced rod and cone length and reduced but not eliminated visual function. However, the developmental phenotypes observed in mice do not match the same severity as the phenotypes observed in humans with Matthew-Wood Syndrome caused by mutant STRA6, suggesting that there might be alternate methods of retinol uptake by the RPE in mice though the majority of retinol uptake is accommodated by STRA6 normally [33]. This study was further supported by research conducted by the Noy lab the following year with a mouse STRA6 knockout where they found that retinoid homeostasis in tissues other than the eye was normal and that the mild loss in visual function from deletion of STRA6 in the mice is due to the high metabolic turnover of vitamin A in the eye without sufficient renewal by alternate retinol uptake methods by the RPE [34].
4.2. Retinol Binding Protein 4 Receptor 2 (RBPR2) in Whole-Body Vitamin A Homeostasis
While STRA6 is expressed in several different organs and tissues, such as the RPE in the eye, it is not expressed in all tissues (Figure 2 and Figure 3). The liver is the main organ involved in the storage of retinoids, however, STRA6 is not expressed in hepatic tissues. Thus, an alternative transport protein is likely expressed in tissues that do not contain STRA6. Discovered by Alapatt and colleagues in 2013, the Retinol Binding Protein 4 Receptor 2 (RBPR2) was found to be the high-affinity RBP4-binding transport protein responsible for the uptake of RBP4- bound retinol in the liver with a similar function as STRA6 in the RPE, however, the efflux capabilities [35]. Publications from our lab showed shown that Rbpr2 was also highly expressed in 11.5 hpf zebrafish embryos at the start of ocular development and in the intestines and pancreas of zebrafish larvae; however, mammalian expression-function patterns of RBPR2 still need further study [36]. The intestinal uptake and efflux of dietary retinol have been theorized to be facilitated by transport proteins alongside passive diffusion, but the specific transport proteins involved were never fully elucidated [37]. Recently, the work from our lab showed that RBPR2 might be involved in the influx (and possible efflux) of retinol in the intestines from the expression patterns of larval zebrafish, though more studies must be performed to establish this function of RBPR2 with mammalian models [36].
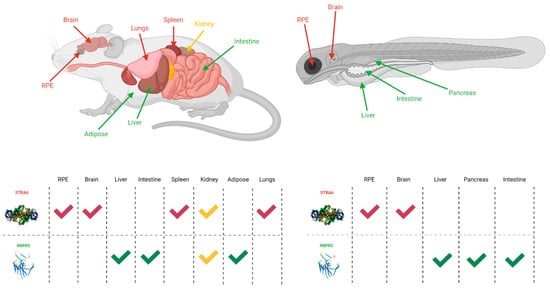
Figure 2. The expression sites of STRA6 and RBPR2 in mouse and zebrafish models [28][32][38][35][36]. Red arrows and checkmarks indicate STRA6 expression in that particular organ, green arrows and checkmarks indicate RBPR2 expression in that particular organ, and yellow arrows and checkmarks indicate expression of both STRA6 and RBPR2 in that organ. STRA6 is consistently expressed in the retinal pigment epithelium, brain, and shares expression with RBPR2 within the kidneys of both organisms. Otherwise, STRA6 and RBPR2 expression is mutually exclusive. Created with BioRender.com.
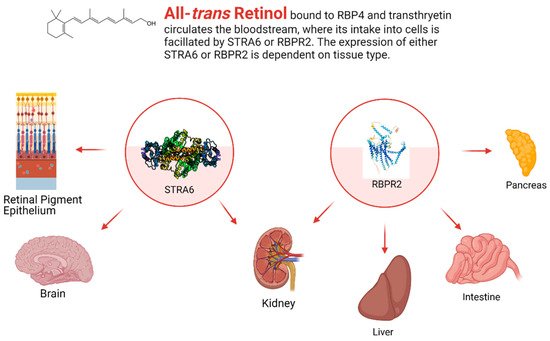
Figure 3. The general expression pattern of STRA6 and RBPR2. Again, STRA6 and RBPR2 share expression within the kidney, with mutually exclusive expression in all other organ systems [28][32][35][36]. Created with BioRender.com.
This entry is adapted from the peer-reviewed paper 10.3390/nu13113987
References
- Borel, P.; Desmarchelier, C. Genetic Variations Associated with Vitamin A Status and Vitamin A Bioavailability. Nutrients 2017, 9, 246.
- Kopec, R.E.; Gleize, B.; Borel, P.; Desmarchelier, C.; Caris-Veyrat, C. Are lutein, lycopene, and β-carotene lost through the digestive process? Food Funct. 2017, 8, 1494–1503.
- Bohn, T.; Desmarchelier, C.; El, S.N.; Keijer, J.; Van Schothorst, E.; Rühl, R.; Borel, P. β-Carotene in the human body: Metabolic bioactivation pathways—From digestion to tissue distribution and excretion. Proc. Nutr. Soc. 2019, 78, 68–87.
- Von Lintig, J.; Moon, J.; Lee, J.; Ramkumar, S. Carotenoid metabolism at the intestinal barrier. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2020, 1865, 158580.
- Von Lintig, J.; Moon, J.; Babino, D. Molecular components affecting ocular carotenoid and retinoid homeostasis. Prog. Retin. Eye Res. 2021, 80, 100864.
- Ramkumar, S.; Moon, J.; Golczak, M.; von Lintig, J. LRAT coordinates the negative-feedback regulation of intestinal retinoid biosynthesis from β-carotene. J. Lipid Res. 2021, 62, 100055.
- Blomhoff, R. Transport and Metabolism of Vitamin A. Nutr. Rev. 2009, 52, S13–S23.
- Chelstowska, S.; Widjaja-Adhi, M.A.K.; Silvaroli, J.A.; Golczak, M. Molecular Basis for Vitamin A Uptake and Storage in Vertebrates. Nutrients 2016, 8, 676.
- Chadwick, A.C.; Jensen, D.R.; Hanson, P.J.; Lange, P.T.; Proudfoot, S.C.; Peterson, F.C.; Volkman, B.F.; Sahoo, D. NMR Structure of the C-Terminal Transmembrane Domain of the HDL Receptor, SR-BI, and a Functionally Relevant Leucine Zipper Motif. Structure 2017, 25, 446–457.
- Valacchi, G.; Sticozzi, C.; Lim, Y.; Pecorelli, A. Scavenger receptor class B type I: A multifunctional receptor. Ann. N. Y. Acad. Sci. 2011, 1229, E1–E7.
- Widjaja-Adhi, M.A.K.; Lobo, G.P.; Golczak, M.; Von Lintig, J. A genetic dissection of intestinal fat-soluble vitamin and carotenoid absorption. Hum. Mol. Genet. 2015, 24, 3206–3219.
- Kiefer, C.; Sumser, E.; Wernet, M.F.; von Lintig, J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc. Natl. Acad. Sci. USA 2002, 99, 10581–10586.
- Lobo, G.P.; Amengual, J.; Palczewski, G.; Babino, D.; von Lintig, J. Mammalian Carotenoid-oxygenases: Key players for carotenoid function and homeostasis. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2012, 1821, 78–87.
- Amengual, J.; Golczak, M.; Palczewski, K.; von Lintig, J. Lecithin:Retinol Acyltransferase Is Critical for Cellular Uptake of Vitamin A from Serum Retinol-binding Protein. J. Biol. Chem. 2012, 287, 24216–24227.
- Von Lintig, J.; Hessel, S.; Isken, A.; Kiefer, C.; Lampert, J.M.; Voolstra, O.; Vogt, K. Towards a better understanding of carotenoid metabolism in animals. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2005, 1740, 122–131.
- Sui, X.; Kiser, P.D.; von Lintig, J.; Palczewski, K. Structural basis of carotenoid cleavage: From bacteria to mammals. Arch. Biochem. Biophys. 2013, 539, 203–213.
- Shete, V.; Quadro, L. Mammalian Metabolism of β-Carotene: Gaps in Knowledge. Nutrients 2013, 5, 4849–4868.
- Lobo, G.P.; Hessel, S.; Eichinger, A.; Noy, N.; Moise, A.R.; Wyss, A.; Palczewski, K.; Von Lintig, J. ISX is a retinoic acid-sensitive gatekeeper that controls intestinal β,β-carotene absorption and vitamin A production. FASEB J. 2010, 24, 1656–1666.
- O’Byrne, S.M.; Blaner, W.S. Retinol and retinyl esters: Biochemistry and physiology. J. Lipid Res. 2013, 54, 1731–1743.
- Blaner, W.S. Vitamin A signaling and homeostasis in obesity, diabetes, and metabolic disorders. Pharmacol. Ther. 2019, 197, 153–178.
- Thompson, S.J.; Sargsyan, A.; Lee, S.-A.; Yuen, J.J.; Cai, J.; Smalling, R.; Ghyselinck, N.; Mark, M.; Blaner, W.S.; Graham, T.E. Hepatocytes Are the Principal Source of Circulating RBP4 in Mice. Diabetes 2016, 66, 58–63.
- Van Bennekum, A.M.; Wei, S.; Gamble, M.V.; Vogel, S.; Piantedosi, R.; Gottesman, M.; Episkopou, V.; Blaner, W.S. Biochemical Basis for Depressed Serum Retinol Levels in Transthyretin-deficient Mice. J. Biol. Chem. 2001, 276, 1107–1113.
- Shirakami, Y.; Lee, S.-A.; Clugston, R.; Blaner, W.S. Hepatic metabolism of retinoids and disease associations. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2012, 1821, 124–136.
- Li, Y.; Wongsiriroj, N.; Blaner, W.S. The multifaceted nature of retinoid transport and metabolism. Hepatobiliary Surg. Nutr. 2014, 3, 126–139.
- Blaner, W.S. STRA6, a Cell-Surface Receptor for Retinol-Binding Protein: The Plot Thickens. Cell Metab. 2007, 5, 164–166.
- Kawaguchi, R.; Yu, J.; Honda, J.; Hu, J.; Whitelegge, J.; Ping, P.; Wiita, P.; Bok, D.; Sun, H. A Membrane Receptor for Retinol Binding Protein Mediates Cellular Uptake of Vitamin A. Science 2007, 315, 820–825.
- Kelly, M.; von Lintig, J. STRA6: Role in Cellular Retinol Uptake and Efflux. Hepatobiliary Surg. Nutr. 2015, 4, 229.
- Bouillet, P.; Sapin, V.; Chazaud, C.; Messaddeq, N.; Décimo, D.; Dollé, P.; Chambon, P. Developmental expression pattern of Stra6, a retinoic acid-responsive gene encoding a new type of membrane protein. Mech. Dev. 1997, 63, 173–186.
- Kawaguchi, R.; Zhong, M.; Kassai, M.; Ter-Stepanian, M.; Sun, H. STRA6-Catalyzed Vitamin A Influx, Efflux, and Exchange. J. Membr. Biol. 2012, 245, 731–745.
- Pasutto, F.; Sticht, H.; Hammersen, G.; Gillessen-Kaesbach, G.; FitzPatrick, D.R.; Nürnberg, G.; Brasch, F.; Schirmer-Zimmermann, H.; Tolmie, J.L.; Chitayat, D.; et al. Mutations in STRA6 Cause a Broad Spectrum of Malformations Including Anophthalmia, Congenital Heart Defects, Diaphragmatic Hernia, Alveolar Capillary Dysplasia, Lung Hypoplasia, and Mental Retardation. Am. J. Hum. Genet. 2007, 80, 550–560.
- Golzio, C.; Martinovic-Bouriel, J.; Thomas, S.; Mougou-Zrelli, S.; Grattagliano-Bessières, B.; Bonnière, M.; Delahaye, S.; Munnich, A.; Encha-Razavi, F.; Lyonnet, S.; et al. Matthew-Wood Syndrome Is Caused by Truncating Mutations in the Retinol-Binding Protein Receptor Gene STRA6. Am. J. Hum. Genet. 2007, 80, 1179–1187.
- Isken, A.; Golczak, M.; Oberhauser, V.; Hunzelmann, S.; Driever, W.; Imanishi, Y.; Palczewski, K.; von Lintig, J. RBP4 Disrupts Vitamin A Uptake Homeostasis in a STRA6-Deficient Animal Model for Matthew-Wood Syndrome. Cell Metab. 2008, 7, 258–268.
- Ruiz, A.; Mark, M.; Jacobs, H.; Klopfenstein, M.; Hu, J.; Lloyd, M.; Habib, S.; Tosha, C.; A Radu, R.; Ghyselinck, N.B.; et al. Retinoid Content, Visual Responses, and Ocular Morphology Are Compromised in the Retinas of Mice Lacking the Retinol-Binding Protein Receptor, STRA6. Investig. Opthalmol. Vis. Sci. 2012, 53, 3027–3039.
- Berry, D.C.; Jacobs, H.; Marwarha, G.; Gely-Pernot, A.; O’Byrne, S.M.; DeSantis, D.; Klopfenstein, M.; Feret, B.; Dennefeld, C.; Blaner, W.S.; et al. The STRA6 Receptor Is Essential for Retinol-binding Protein-induced Insulin Resistance but Not for Maintaining Vitamin A Homeostasis in Tissues Other Than the Eye. J. Biol. Chem. 2013, 288, 24528–24539.
- Alapatt, P.; Guo, F.; Komanetsky, S.M.; Wang, S.; Cai, J.; Sargsyan, A.; Diaz, E.; Bacon, B.; Aryal, P.; Graham, T.E. Liver Retinol Transporter and Receptor for Serum Retinol-binding Protein (RBP4). J. Biol. Chem. 2013, 288, 1250–1265.
- Shi, Y.; Obert, E.; Rahman, B.; Rohrer, B.; Lobo, G.P. The Retinol Binding Protein Receptor 2 (Rbpr2) is required for Photoreceptor Outer Segment Morphogenesis and Visual Function in Zebrafish. Sci. Rep. 2017, 7, 16207.
- During, A.; Harrison, E.H. Mechanisms of provitamin A (carotenoid) and vitamin A (retinol) transport into and out of intestinal Caco-2 cells. J. Lipid Res. 2007, 48, 2283–2294.
- Amengual, J.; Zhang, N.; Kemerer, M.; Maeda, T.; Palczewski, K.; von Lintig, J. STRA6 is critical for cellular vitamin A uptake and homeostasis. Hum. Mol. Genet. 2014, 23, 5402–5417.
This entry is offline, you can click here to edit this entry!
