All biological processes associated with high sports performance, including energy metabolism, are influenced by genetics. DNA sequence variations in such genes, single nucleotide variants (SNVs), could confer genetic advantages that can be exploited to achieve optimal athletic performance. Ignorance of these features can create genetic “barriers” that prevent professional athletes from pursuing a career in sports.
- personalized medicine
- sports genetics
- candidate genes
- single nucleotide variant
- polymorphism
- energy metabolism
- skeletal muscles
- athlete
1. Introduction
The proportion of competitive period in the annual training cycle in cyclic sports (such as running disciplines of athletics) has significantly increased [1]. This makes high demands on the physiological and biochemical aspects of training athletes on the first place. It should be noted that running disciplines vary from sprint, which last for a few seconds, to marathon taking hours to run [2]. Accordingly, the competitive result in cyclic sports will be determined, firstly, by the availability of adenosine triphosphate (ATP) as the main substrate of energy supply and, secondly, by the number of skeletal muscle motor units involved. As presented in Figure 1 , the energy supply for muscle activity is provided by three energy systems: phosphogenic pathway, glycolytic pathway, and mitochondrial respiration ( Figure 1 ).
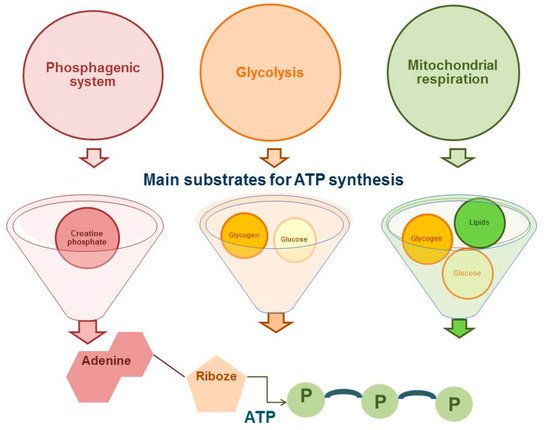
All energy systems for the synthesis of ATP molecules use a different substrate, involve a different number of reactions, resulting in having a different regeneration rate of ATP molecules and different metabolic products, which will determine the contribution of each system to the process of fatigue of the athlete’s skeletal muscles and a movement speed decrease, which is important in cyclic sports [3][4][5].
Thus, the energy supply of muscular activity and the ratio of different types of fibers in skeletal muscles are important factors determining an athlete’s performance in cyclic sports. Consequently, the identification of genetic markers, single nucleotide variants (SNVs), associated with the metabolic and contractile efficiency of skeletal muscles is a priority area of sports genetics. The identification of an athlete’s predisposition to better tolerance of anaerobic or aerobic loads contributes to the rational choice of sports loads and the prevention of sports injuries [5].
Objective of the thematic review—to conduct a research on candidate genes associated with regulation of skeletal muscle energy metabolism in athletes.
2. Development and Findings
According to our analysis of studies of candidate genes encoding structural proteins and enzymes involved in the regulation of energy metabolism in skeletal muscles, the researchers’ interest in sports genetics has been increasing in recent years. The most studied are 6 candidate genes ( Table 1 ), the expression level of which differs in skeletal muscles, myocardium, and lungs ( Table 2 ), which is important to consider when translating the results of genetic research into real sports practice.
| Gene | Localization, Chromosome | Protein/ Enzyme |
Effects on Energy Metabolism of Skeletal Muscle |
|---|---|---|---|
| AMPD1 | 1p13.2 | Adenosine monophosphate deaminase 1 type (AMPD1) | AMPD1 catalyzes the deamination of adenosine monophosphate and inosine monophosphate in skeletal muscle and plays an important role in the purine nucleotide cycle. |
| PPARG | 3p25.2 | Peroxisome proliferator activated receptor gamma (PPARG) | PPARG controls the peroxisome beta-oxidation pathway of fatty acids and is a key regulator of adipocyte differentiation and glucose homeostasis. |
| PPARGC1A | 4p15.2 | Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1 Alpha (PPARGC1A) |
PPARGC1A regulates genes involved in energy metabolism, provides a direct link between external physiological stimuli and regulation of mitochondrial biogenesis, and is the main factor regulating the determination of muscle fiber type. |
| PPARA | 22q13.31 | Peroxisome Proliferator Activated Receptor Alpha (PPARA) |
PPARA is involved in the regulation of energy metabolism, regulates the expression of genes encoding several key muscle enzymes involved in fatty acid oxidation. |
| CKM | 19q13.32 | Creatine Kinase, Muscle (CKM) |
CKM catalyzes the transfer of phosphate between ATP and various phosphogenic groups such as creatine phosphate; CKM isozymes play a central role in energy transduction in tissues with high energy requirements such as skeletal muscle, heart, brain. |
| TFAM | 10q21.1 | Transcription Factor A, Mitochondrial (TFAM) |
TFAM is responsible for regulating mitochondrial DNA replication and transcription and also protects cells from oxidative stress. |
| Gene | Expression Level in Skeletal Muscles (RPKM) |
Expression Level in Myocardium (RPKM) |
Expression Level in Lung (RPKM) |
|---|---|---|---|
| AMPD1 | 225.7 | 0.023 | 0.363 |
| PPARG | 2.097 | 4.716 | 19.45 |
| PPARGC1A | 11.02 | 9.406 | 3.292 |
| PPARA | 12.34 | 7.889 | 5.624 |
| CKM | 25890.0 | 2987 | 4.667 |
| TFAM | 6.0 | 4.754 | 9.471 |
High expression of PPARGC1A is noted in metabolically active tissues with many mitochondria and oxidative phosphorylation, such as heart and skeletal muscle ( Table 2 ).
SNVs are described in the coding region of the PPARGC1A gene, which are associated with muscle energy metabolism. SNV C23815662T (rs8192678) leads to the replacement of glycine with serine (Gly482Ser), which has a functional significance in the adaptation of skeletal muscles to physical stress [8][9][10][11]. There is evidence that this SNV is associated with changes in blood lipids and insulin sensitivity. Carriers of Ser have a higher level of low density lipoproteins in the blood serum and higher insulin resistance compared to carriers of Gly, as well as reduced energy metabolism of skeletal muscles [12][13].
Studies have shown that the PPARGC1A gene plays an important part in maintaining the expression of mitochondrial metabolic and antioxidant enzymes in skeletal muscles and affects training-induced adaptation of mitochondrial proteins in skeletal muscles. Regular aerobic exercises increase the expression/activity of membrane transporters and mitochondrial metabolic enzymes, and increase capillarization in skeletal muscles, altogether increasing the oxidative capacity of muscle fibres and the ability of myocytes to oxidize carbohydrates and fatty acids. This increases the expression of antioxidant defense enzymes [14], potentially providing better protection against reactive oxygen species in skeletal muscles.
3. Conclusions
Possible, identification of genetic biomarkers associated with the regulation of energy metabolism in skeletal muscles in athletes may help sports physicians and coaches develop personalized strategies for selecting children, teenagers and young adults for endurance, strength and speed sports. However, the multifactorial aspect of sport performances, including impact of genetics, epigenetics, environment (training, resting, nutrition, psycho-emotional status and etc.), is important for personalized strategies for selecting of athletes. This approach could improve sports performance and reduce the risk of sports injuries to the musculoskeletal system.
This entry is adapted from the peer-reviewed paper 10.3390/genes12111682
References
- Balberova, O.V.; Sidorkina, E.G.; Koshkina, K.S.; Plachy, J.K.; Bykov, E.V. Model characteristics of competition performance in terms of atletes, functional fitness. Sci. Educ. Today 2021, 3, 161–176.
- Kryazhev, V.D.; Kryazheva, S.V.; Alenurov, E.A.; Bokova, L.V. Competitive and training areas in cyclical locomotion at top-qualified athletes. Sci. Lett. P.F. Lesgaft Univ. 2020, 10, 205–213.
- Hargreaves, M.; Spriet, L.L. Skeletal muscle energy metabolism during exercise. Nat. Metab. 2020, 2, 817–828.
- Baker, S.; Mc Cormick, M.C.; Robergs, R.A. Interaction among Skeletal Muscle Metabolic Energy Systems during Intense Exercise. J. Nutr. Metab. 2010, 2010, 905612.
- Balberova, O.V. Candidate genes and single-nucleotide gene variants associated with muscle and tendon injuries in cyclic sports athletes. Pers. Psychiatry Neurol. 2021, 1, 64–72.
- GeneCards: The Human Gene Database. Available online: https://www.genecards.org/ (accessed on 20 August 2021).
- GTEx Portal. Available online: https://www.gtexportal.org/ (accessed on 28 August 2021).
- Tharabenjasin, P.; Pabalan, N.; Jarjanazi, H. Association of PPARGC1A Gly428Ser (rs8192678) polymorphism with potential for athletic ability and sports performance: A meta-analysis. PLoS ONE 2019, 14, e0200967.
- Handschin, C.; Spiegelman, B.M. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature 2008, 454, 463–469.
- Ahmetov, I.I.; Mozhayskaya, I.A.; Flavell, D.M.; Astratenkova, I.V.; Komkova, A.I.; Lyubaeva, E.V.; Tarakin, P.P.; Shenkman, B.S.; Vdovina, A.B.; Netreba, A.I.; et al. PPARalpha gene variation and physical performance in Russian athletes. Eur. J. Appl. Physiol. 2006, 97, 103–108.
- Mathai, A.S.; Bonen, A.; Benton, C.R.; Robinson, D.L.; Graham, T.E. Rapid exercise-induced changes in PGC-1alpha mRNA and protein in human skeletal muscle. J. Appl. Physiol. 2008, 105, 1098–1105.
- Sharma, R.; Matharoo, K.; Kapoor, R.; Bhanwer, A.J.S. Association of PGC-1α gene with type 2 diabetes in three unrelated endogamous groups of North-West India (Punjab): A case-control and meta-analysis study. Mol. Genet. Genom. 2018, 293, 317–329.
- Yang, Y.; Mo, X.; Chen, S.; Lu, X.; Gu, D. Association of peroxisome proliferator-activated receptor gamma coactivator 1 Alpha (PPARGC1A) gene polymorphisms and type 2 diabetes mellitus: A meta-analysis. Diabetes Metab. Res. Rev. 2011, 27, 177–184.
- Ilyutik, A.; Gilep, I. The relationship of gene polymorphisms with the development of physical qualities in athletes (based on the material of speed skating). Sci. Olymp. Sports 2017, 3, 51–57. (In Russian)
