microRNAs (miRNAs) have been documented to be regulators of valvular diseases pathogenesis, diagnostic biomarkers, and therapeutical targets. They in fact play stimulatory or inhibitory roles in mitral valve prolapse development, aortic leaflet fusion, and calcification pathways. Tissue expression assessment and comparison between physiological and pathological phenotypes of different disease entities, including mitral valve prolapse and mitral chordae tendineae rupture, emerged as the best strategies to address miRNAs over or under-representation and thus, their impact on pathogeneses.
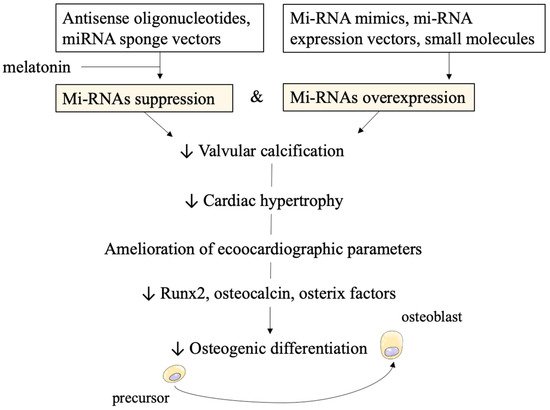
miRNAs can also be targeted by several molecules. Inhibitors such as antisense oligonucleotides and sponge vectors are under investigation. Furthermore, to increase miRNAs activity miRNA mimics, miRNA expression vectors, and small molecules have been developed.
- miRNAs
- valvular heart diseases
- aortic stenosis
- calcification
- mitral valve prolapse
- aortic valve defect
- vectors
- delivery systems
- nanoparticles
1. miRNAs Effects on Aortic Stenosis and Mitral Prolapse Necessity of Novel Biomarkers
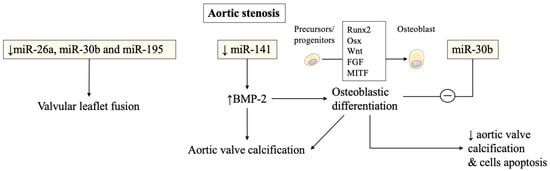
In aortic stenosis, miRNAs were also found to be altered in stenotic aortic valve leaflets compared to insufficient ones by Nigam et al. [1]. The reduced levels of miR-26a, miR-30b, and miR-195 reported in stenotic valves, were almost certainly responsible for their higher risk of developing valvular leaflet fusion. The change of morphology is due to accelerated calcium accumulation compared to regurgitant aortic valves without morphological fusion of valve leaflets. These miRNAs were involved in the biological processes that modulate calcification-related genes in vitro [1]. Other studies have confirmed miRNAs’ involvement in calcification pathways and will be discussed later.
Several studies have then recorded an important association of miRNAs with post-transcriptional regulation of gene expression in aortic valve stenosis. miR-141 was found to be a regulator of the levels of bone morphogenetic protein 2 (BMP-2), whereby unrestrained activity led to calcification of the aortic valve mediated by a stimulation of osteogenesis. miR-141 was markedly attenuated in patients with aortic stenosis associated with the bicuspid aortic valve [2]. Yanagawa et al. proposed this new key role of miR-141 in the modulation of aortic valve calcification disorders, highlighting the strategic therapeutic target that emerged in the assessment of progressive calcification in stenotic aortic valve disease [1][2]. The peculiar morphologic features of the stenotic aortic valve may probably be explained by the lower expression of miR-30b which is a known repressor of bone morphogenetic protein 2-mediated osteogenesis [2].
Zhang et al. demonstrated the role of miR-30b in reducing osteoblast differentiation activity induced by bone morphogenetic protein 2 [3]. The latter was implicated in promoting calcific aortic valve disease. The expression of miR-30b was found effective in reducing the risk of human aortic valve calcification and apoptosis through direct targeting of Runx2, Smad1, and caspase-3 [3] ( Figure 1 ).
Plasma levels of miRNAs have been used to monitor degenerative disease of the mitral valve (MV) but have not been widely adopted by the cardiology community [4][1][2][5][6][7]. The lack of enthusiasm of cardiologists for investigations into degenerative aortic valve stenosis [1][2][3][8][9] can be partially deduced from the lack of positive findings in mitral valve degenerative diseases leading to regurgitation [10][11][12][13][14][15].
2. Altered miRNAs Expression in Congenital Valve Disorders and Cardiogenetic Processes
3. Novel Therapeutical Strategies: miRNAs Targeting to Suppress or Activate Them
3.1. Results from In-Vivo and In-Vitro Testing for Aortic Valvular Stenosis
3.2. Technical Concerns on Stability and Efficacy of miRNAs as Therapeutical Targets
3.3. Disadvantages in Pharmacokinetics and Proposed Mechanisms for Delivery Vehicles
This entry is adapted from the peer-reviewed paper 10.3390/ijms222212132
References
- Nigam, V.; Sievers, H.H.; Jensen, B.C.; Sier, H.A.; Simpson, P.C.; Srivastava, D.; Mohamed, S.A. Altered microRNAs in bicuspid aortic valve: A comparison between stenotic and insufficient valves. J. Heart Valve Dis. 2010, 19, 459–465.
- Yanagawa, B.; Lovren, F.; Pan, Y.; Garg, V.; Quan, A.; Tang, G.; Singh, K.K.; Shukla, P.C.; Kalra, N.P.; Peterson, M.D.; et al. miRNA-141 is a novel regulator of BMP-2-mediated calcification in aortic stenosis. J. Thorac. Cardiovasc. Surg. 2012, 144, 256–406.
- Zhang, M.; Liu, X.; Zhang, X.; Song, Z.; Han, L.; He, Y.; Xu, Z. MicroRNA-30b is a multifunctional regulator of aortic valve interstitial cells. J. Thorac. Cardiovasc. Surg. 2014, 147, 1073–1080.e2.
- Mayeux, R. Biomarkers: Potential uses and limitations. NeuroRX 2004, 1, 182–188.
- Song, R.; Fullerton, D.A.; Ao, L.; Zhao, K.; Reece, T.B.; Cleveland, J.C.; Meng, X. Altered MicroRNA Expression Is Responsible for the Pro-Osteogenic Phenotype of Interstitial Cells in Calcified Human Aortic Valves. J. Am. Hear. Assoc. 2017, 6, e005364.
- Kumar, D.; Narang, R.; Sreenivas, V.; Rastogi, V.; Bhatia, J.; Saluja, D.; Srivastava, K. Circulatory miR-133b and miR-21 as Novel Biomarkers in Early Prediction and Diagnosis of Coronary Artery Disease. Genes 2020, 11, 164.
- Ikeda, S.; Kong, S.W.; Lu, J.; Bisping, E.; Zhang, H.; Allen, P.D.; Golub, T.R.; Pieske, B.; Pu, W.T. Altered microRNA expression in human heart disease. Physiol. Genom. 2007, 31, 367–373.
- Guo, X.; Chen, Y.; Lu, Y.; Li, P.; Yu, H.; Diao, F.-R.; Tang, W.-D.; Hou, P.; Zhao, X.-X.; Shi, C.-Y. High level of circulating microRNA-142 is associated with acute myocardial infarction and reduced survival. Ir. J. Med. Sci. 2020, 189, 933–937.
- Sabatino, J.; Wicik, Z.; De Rosa, S.; Eyileten, C.; Jakubik, D.; Spaccarotella, C.; Mongiardo, A.; Postula, M.; Indolfi, C. MicroRNAs fingerprint of bicuspid aortic valve. J. Mol. Cell. Cardiol. 2019, 134, 98–106.
- Songia, P.; Branchetti, E.; Parolari, A.; Myasoedova, V.; Ferrari, G.; Alamanni, F.; Tremoli, E.; Poggio, P. Mitral valve endothelial cells secrete osteoprotegerin during endothelial mesenchymal transition. J. Mol. Cell. Cardiol. 2016, 98, 48–57, 386–387.
- Songia, P.; Porro, B.; Chiesa, M.; Myasoedova, V.; Alamanni, F.; Tremoli, E.; Poggio, P. Identification of Patients Affected by Mitral Valve Prolapse with Severe Regurgitation: A Multivariable Regression Model. Oxidative Med. Cell. Longev. 2017, 2017, 1–6.
- Tan, H.T.; Ling, L.H.; Dolor-Torres, M.C.; Yip, J.W.-L.; Richards, A.M.; Chung, M.C. Proteomics discovery of biomarkers for mitral regurgitation caused by mitral valve prolapse. J. Proteom. 2013, 94, 337–345.
- Hulanicka, M.; Garncarz, M.; Parzeniecka-Jaworska, M.; Jank, M. Plasma miRNAs as potential biomarkers of chronic degenerative valvular disease in Dachshunds. BMC Veter. Res. 2014, 10, 205.
- Li, Q.; Freeman, L.M.; Rush, J.E.; Laflamme, D.P. Expression Profiling of Circulating MicroRNAs in Canine Myxomatous 410 Mitral Valve Disease. Int. J. Mol. Sci. 2015, 16, 14098–14108.
- Bulent Vatan, M.; Kalayci Yigin, A.; Akdemir, R.; Tarik Agac, M.; Akif Cakar, M.; Aksoy, M.; Tatli, E.; Kilic, H.; Gunduz, H.; Guzel, D.; et al. Altered Plasma MicroRNA Expression in Patients with Mitral Chordae Tendineae Rupture. J. Heart Valve Dis. 2016, 25, 580–588.
- Sun, R.; Liu, M.; Lu, L.; Zheng, Y.; Zhang, P. Congenital Heart Disease: Causes, Diagnosis, Symptoms, and Treatments. Cell Biophys. 2015, 72, 857–860.
- Dolbec, K.; Mick, N.W. Congenital heart disease. Emerg. Med. Clin. N. Am. 2011, 29, 811–827.
- Higgins, S.S.; Reid, A. Common congenital heart defects. Long-term follow-up. Nurs. Clin. N. Am. 1994, 29, 233–248.
- Chen, J.-F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.-Z. The Role of MicroRNA-1 and MicroRNA-133 in Skeletal Muscle Proliferation and Differentiation. Nat. Genet. 2005, 38, 228–233.
- Yin, V.P.; Lepilina, A.; Smith, A.; Poss, K.D. Regulation of zebrafish heart regeneration by miR-133. Dev. Biol. 2012, 365, 319–327.
- Porrello, E.R.; Mahmoud, A.I.; Simpson, E.; Johnson, B.A.; Grinsfelder, D.; Canseco, D.; Mammen, P.P.; Rothermel, B.A.; Olson, E.N.; Sadek, H.A. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl. Acad. Sci. USA 2013, 110, 187–192.
- Eulalio, A.; Mano, M.; Ferro, M.D.; Zentilin, L.; Sinagra, G.; Zacchigna, S.; Giacca, M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 2012, 492, 376–381.
- Wojciechowska, A.; Braniewska, A.; Kozar-Kamińska, K. MicroRNA in cardiovascular biology and disease. Adv. Clin. Exp. Med. 2017, 26, 868–874.
- Yoshida, T.; Delafontaine, P. Mechanisms of IGF-1-Mediated Regulation of Skeletal Muscle Hypertrophy and Atrophy. Cells 2020, 9, 1970.
- Zhou, H.; Wang, B.; Yang, Y.-X.; Jia, Q.-J.; Zhang, A.; Qi, Z.-W.; Zhang, J.-P. Long Noncoding RNAs in Pathological Cardiac Remodeling: A Review of the Update Literature. BioMed Res. Int. 2019, 2019, 1–11.
- Simmons, C.A.; Zilberberg, J.; Davies, P.F. A rapid, reliable method to isolate high quality endothelial RNA from small spatially-defined locations. Ann. Biomed. Eng. 2004, 32, 1453–1459.
- Simmons, C.A.; Grant, G.; Manduchi, E.; Davies, P. Spatial Heterogeneity of Endothelial Phenotypes Correlates With Side-Specific Vulnerability to Calcification in Normal Porcine Aortic Valves. Circ. Res. 2005, 96, 792–799.
- Scolari, F.L.; Faganello, L.S.; Garbin, H.I.; e Mattos, B.P.; Biolo, A. A systematic review of microRNAs in patients with hypertrophic cardiomyopathy. Int. J. Cardiol. 2020, 327, 146–154.
- Villar, A.V.; Merino, D.; Wenner, M.; Llano, M.; Cobo, M.; Montalvo, C.; García, R.; Martín-Durán, R.; Hurlé, J.M.; A Hurlé, M.; et al. Myocardial gene expression of microRNA-133a and myosin heavy and light chains, in conjunction with clinical parameters, predict regression of left ventricular hypertrophy after valve replacement in patients with aortic stenosis. Heart 2011, 97, 1132–1137.
- García, R.; Villar, A.V.; Cobo, M.; Llano, M.; Martín-Durán, R.; Hurlé, M.A.; Nistal, J.F. Circulating Levels of miR-133a Predict the Regression Potential of Left Ventricular Hypertrophy After Valve Replacement Surgery in Patients With Aortic Stenosis. J. Am. Hear. Assoc. 2013, 2, e000211.
- Dixit, G.; Schanz, W.; Pappas, B.; Maretzky, T. Members of the Fibroblast Growth Factor Receptor Superfamily Are Proteolytically Cleaved by Two Differently Activated Metalloproteases. Int. J. Mol. Sci. 2021, 22, 3165.
- Rathan, S.; Ankeny, C.J.; Arjunon, S.; Ferdous, Z.; Kumar, S.; Esmerats, J.F.; Heath, J.M.; Nerem, R.M.; Yoganathan, A.P.; Jo, H. Identification of side- and shear-dependent microRNAs regulating porcine aortic valve pathogenesis. Sci. Rep. 2016, 6, 25397.
- Heath, J.M.; Esmerats, J.F.; Khambouneheuang, L.; Kumar, S.; Simmons, R.; Jo, H. Mechanosensitive microRNA-181b Regulates Aortic Valve Endothelial Matrix Degradation by Targeting TIMP3. Cardiovasc. Eng. Technol. 2017, 9, 141–150.
- Sun, X.; Icli, B.; Wara, A.K.; Belkin, N.; He, S.; Kobzik, L.; Hunninghake, G.M.; Pinilla-Vera, M.; Blackwell, T.S.; Baron, R.M.; et al. MicroRNA-181b regulates NF-κB-mediated vascular inflammation. J. Clin. Investig. 2012, 122, 1973–1990.
- Larivière, R.; Gauthier-Bastien, A.; Ung, R.-V.; St-Hilaire, J.; Mac-Way, F.; Richard, D.; Agharazii, M. Endothelin type A receptor blockade reduces vascular calcification and inflammation in rats with chronic kidney disease. J. Hypertens. 2017, 35, 376–384.
- Wu, S.Y.; Zhang, B.H.; Pan, C.S.; Jiang, H.F.; Pang, Y.Z.; Tang, C.S.; Qi, Y.F. Endothelin-1 is a potent regulator in vivo in vascular calcification and in vitro in calcification of vascular smooth muscle cells. Peptides 2003, 24, 1149–1156.
- Ryu, J.; Kwon, D.-H.; Choe, N.; Shin, S.; Jeong, G.; Lim, Y.-H.; Kim, J.; Park, W.J.; Kook, H.; Kim, Y.-K. Characterization of Circular RNAs in Vascular Smooth Muscle Cells with Vascular Calcification. Mol. Ther.-Nucleic Acids 2019, 19, 31–41.
- Henein, M.; Granåsen, G.; Wiklund, U.; Schmermund, A.; Guerci, A.; Erbel, R.; Raggi, P. High dose and long-term statin therapy accelerate coronary artery calcification. Int. J. Cardiol. 2015, 184, 581–586.
- Toshima, T.; Watanabe, T.; Narumi, T.; Otaki, Y.; Shishido, T.; Aono, T.; Goto, J.; Watanabe, K.; Sugai, T.; Takahashi, T.; et al. Therapeutic inhibition of microRNA-34a ameliorates aortic valve calcification via modulation of Notch1-Runx2 signaling. Cardiovasc. Res. 2019, 116, 983–994.
- Wang, Y.; Han, D.; Zhou, T.; Zhang, J.; Liu, C.; Cao, F.; Dong, N. Melatonin ameliorates aortic valve calcification via the regulation of circular RNA CircRIC3/miR-204-5p/DPP4 signaling in valvular interstitial cells. J. Pineal. Res. 2020, 69, e12666.
- Van der Ven, C.F.; Wu, P.J.; Tibbitt, M.W.; van Mil, A.; Sluijter, J.P.; Langer, R.; Aikawa, E. In vitro 3D model and miRNA drug delivery to target calcific aortic valve disease. Clin. Sci. 2017, 131, 181–195.
- Ebert, M.S.; Neilson, J.R.; A Sharp, P. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726.
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008, 456, 980–984.
- Ma, L.; Reinhardt, F.; Pan, E.; Soutschek, J.; Bhat, B.; Marcusson, E.G.; Teruya-Feldstein, J.; Bell, G.W.; A Weinberg, R. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010, 28, 341–347.
- Davis, S.; Lollo, B.; Freier, S.; Esau, C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006, 34, 2294–2304.
- Alexis, F.; Pridgen, E.; Molnar, L.K.; Farokhzad, O.C. Factors Affecting the Clearance and Biodistribution of Polymeric Nanoparticles. Mol. Pharm. 2008, 5, 505–515.
- Kanasty, R.L.; A Whitehead, K.; Vegas, A.J.; Anderson, D.G. Action and Reaction: The Biological Response to siRNA and Its Delivery Vehicles. Mol. Ther. 2012, 20, 513–524.
- Whitehead, K.A.; Langer, R. Anderson DG Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 2009, 8, 129–138.
- Tafer, H.; Ameres, S.L.; Obernosterer, G.; A Gebeshuber, C.; Schroeder, R.; Martinez, J.; Hofacker, I. The impact of target site accessibility on the design of effective siRNAs. Nat. Biotechnol. 2008, 26, 578–583.
- Davis, M.E.; Chen, Z.; Shin, D.M. Nanoparticle therapeutics: An emerging treatment modality for cancer. Nat. Rev. Drug Discov. 2008, 7, 771–782.
- Chen, M.Y.; Hoffer, A.; Morrison, P.F.; Hamilton, J.F.; Hughes, J.; Schlageter, K.S.; Lee, J.; Kelly, B.R.; Oldfield, E.H. Surface properties, more than size, limiting convective distribution of virus-sized particles and viruses in the central nervous system. J. Neurosurg. 2005, 103, 311–319.
- Bartlett, D.W.; Davis, M.E. Physicochemical and Biological Characterization of Targeted, Nucleic Acid-Containing Nanoparticles. Bioconjugate Chem. 2007, 18, 456–468.
- Hong, S.; Leroueil, P.R.; Majoros, I.J.; Orr, B.G.; Baker, J.R.; Holl, M.M.B. The Binding Avidity of a Nanoparticle-Based Multivalent Targeted Drug Delivery Platform. Chem. Biol. 2007, 14, 107–115.
- Malam, Y.; Loizidou, M.; Seifalian, A.M. Liposomes and nanoparticles: Nanosized vehicles for drug delivery in cancer. Trends Pharmacol. Sci. 2009, 30, 592–599.
- Allen, T.M.; Cullis, P.R. Liposomal drug delivery systems: From concept to clinical applications. Adv. Drug Deliv. Rev. 2013, 65, 36–48.
- Abu Lila, A.S.; Ishida, T. Liposomal Delivery Systems: Design Optimization and Current Applications. Biol. Pharm. Bull. 2017, 40, 1–10.
- Barile, L.; Vassalli, G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 2017, 174, 63–78.
- Van Rooij, T.; Luan, Y.; Renaud, G.; van der Steen, A.F.; Versluis, M.; de Jong, N.; Kooiman, K. Non-linear response and viscoelastic properties of lipid-coated microbubbles: DSPC versus DPPC. Ultrasound Med. Biol. 2015, 41, 1432–1445.
- Langeveld, S.; Beekers, I.; Collado-Lara, G.; van der Steen, A.; de Jong, N.; Kooiman, K. The Impact of Lipid Handling and Phase Distribution on the Acoustic Behavior of Microbubbles. Pharmaceutics 2021, 13, 119.
- Daeichin, V.; van Rooij, T.; Skachkov, I.; Ergin, B.; Specht, P.A.; Lima, A.; Ince, C.; Bosch, J.G.; van der Steen, A.F.; de Jong, N.; et al. Microbubble Composition and Preparation for High-Frequency Contrast-Enhanced Ultrasound Imaging: In Vitro and In Vivo Evaluation. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2017, 64, 555–567.
- Upadhyay, A.; Dalvi, S.V.; Gupta, G.; Khanna, N. Effect of PEGylation on performance of protein microbubbles and its comparison with lipid microbubbles. Mater. Sci. Eng. C 2017, 71, 425–430.
- Gilam, A.; Conde, J.; Weissglas-Volkov, D.; Oliva-Jorge, N.; Friedman, E.; Artzi, N.; Shomron, N. Local microRNA delivery targets Palladin and prevents metastatic breast cancer. Nat. Commun. 2016, 7, 12868.
- Conde, J.; Oliva-Jorge, N.; Artzi, N. Implantable hydrogel embedded dark-gold nanoswitch as a theranostic probe to sense and overcome cancer multidrug resistance. Proc. Natl. Acad. Sci. USA 2015, 112, E1278–E1287.
- Segovia, N.; Pont, M.; Oliva, N.; Ramos, V.; Borrós, S.; Artzi, N. Hydrogel Doped with Nanoparticles for Local Sustained Release of siRNA in Breast Cancer. Adv. Heal. Mater. 2014, 4, 271–280.
- Conde, J.; Oliva-Jorge, N.; Atilano, M.; Song, H.S.; Artzi, N. Self-assembled RNA-triple-helix hydrogel scaffold for microRNA modulation in the tumour microenvironment. Nat. Mater. 2015, 15, 353–363.
