Cystic Fibrosis (CF) is one of the most common genetic diseases prevalent among the Caucasian population and is caused by mutations of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. To date, several hundred disease-causing mutations are known, resulting in a vast range of geno- and phenotypes, which makes the development of therapeutics especially challenging. To support the development of novel therapeutics, systems biological disease maps can be used. Disease maps represent existing knowledge on disease mechanisms in a computationally readable and comprehensive manner so they can then be used by clinicians and experimental scientists as well as computational scientists for different purposes, such as structuring high-throughput data, identifying disease biomarkers, developing better diagnostics and also identifying potential drug targets and drug repositioning. The CFTR Lifecycle Map in particular details the biogenesis of CFTR in cells to support ongoing drug discovery endeavours in CF research.
- cystic fibrosis
- CFTR
- CFTR maturation
- systems medicine model
- trafficking
- CFTR modulators
1. Introduction
2. CFTR Map
2.1. Representation of the CFTR Lifecycle in the CFTR Core Map
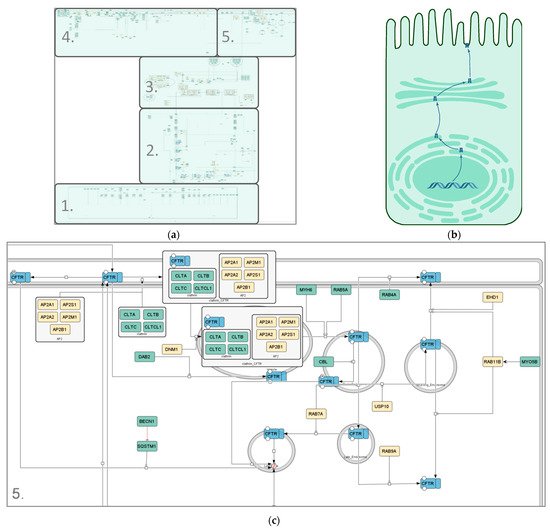
3.1. Visualization of the wt-CFTR Interactome as Coarse Model
4. Conclusion
This entry is adapted from the peer-reviewed paper 10.3390/ijms22147590
References
- Bobadilla, J.L.; Macek, M.; Fine, J.P.; Farrell, P.M. Cystic fibrosis: A worldwide analysis of CFTR mutations—Correlation with incidence data and application to screening. Hum. Mutat. 2002, 19, 575–606.
- O’Sullivan, B.P.; Freedman, S.D. Cystic fibrosis. Lancet 2009, 373, 1891–1904.
- Elborn, J.S. Cystic fibrosis. Lancet 2016, 388, 2519–2531.
- Cystic Fibrosis Mutation Database. Available online: http://www.genet.sickkids.on.ca/ (accessed on 26 January 2021).
- Welcome to CFTR2 | CFTR2. Available online: https://www.cftr2.org/ (accessed on 26 January 2021).
- Sosnay, P.R.; Siklosi, K.R.; Van Goor, F.; Kaniecki, K.; Yu, H.; Sharma, N.; Ramalho, A.S.; Amaral, M.D.; Dorfman, R.; Zielenski, J.; et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat. Genet. 2013, 45, 1160–1167.
- Martiniano, S.L.; Sagel, S.D.; Zemanick, E.T. Cystic fibrosis: A model system for precision medicine. Curr. Opin. Pediatr. 2016, 28, 312–317.
- Southern, K.W.; Patel, S.; Sinha, I.P.; Nevitt, S.J. Correctors (specific therapies for class II CFTR mutations) for cystic fibrosis. Cochrane Database Syst. Rev. 2018.
- Pedemonte, N.; Lukacs, G.L.; Du, K.; Caci, E.; Zegarra-Moran, O.; Galietta, L.J.V.; Verkman, A.S. Small-molecule correctors of defective ΔF508-CFTR cellular processing identified by high-throughput screening. J. Clin. Investig. 2005.
- Van Goor, F.; Hadida, S.; Grootenhuis, P.D.J.; Burton, B.; Cao, D.; Neuberger, T.; Turnbull, A.; Singh, A.; Joubran, J.; Hazlewood, A.; et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc. Natl. Acad. Sci. USA 2009.
- Berg, A.; Hallowell, S.; Tibbetts, M.; Beasley, C.; Brown-Phillips, T.; Healy, A.; Pustilnik, L.; Doyonnas, R.; Pregel, M. High-Throughput Surface Liquid Absorption and Secretion Assays to Identify F508del CFTR Correctors Using Patient Primary Airway Epithelial Cultures. SLAS Discov. 2019.
- De Wilde, G.; Gees, M.; Musch, S.; Verdonck, K.; Jans, M.; Wesse, A.S.; Singh, A.K.; Hwang, T.C.; Christophe, T.; Pizzonero, M.; et al. Identification of GLPG/ABBV-2737, a novel class of corrector, which exerts functional synergy with other CFTR modulators. Front. Pharmacol. 2019, 10.
- Merkert, S.; Schubert, M.; Olmer, R.; Engels, L.; Radetzki, S.; Veltman, M.; Scholte, B.J.; Zöllner, J.; Pedemonte, N.; Galietta, L.J.V.; et al. High-Throughput Screening for Modulators of CFTR Activity Based on Genetically Engineered Cystic Fibrosis Disease-Specific iPSCs. Stem Cell Reports 2019.
- Van Goor, F.; Hadida, S.; Grootenhuis, P.D.J.; Burton, B.; Stack, J.H.; Straley, K.S.; Decker, C.J.; Miller, M.; McCartney, J.; Olson, E.R.; et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. USA 2011.
- Phuan, P.W.; Veit, G.; Tan, J.A.; Finkbeiner, W.E.; Lukacs, G.L.; Verkman, A.S. Potentiators of defective DF508-CFTR gating that do not interfere with corrector action. Mol. Pharmacol. 2015.
- Carlile, G.W.; Robert, R.; Goepp, J.; Matthes, E.; Liao, J.; Kus, B.; Macknight, S.D.; Rotin, D.; Hanrahan, J.W.; Thomas, D.Y. Ibuprofen rescues mutant cystic fibrosis transmembrane conductance regulator trafficking. J. Cyst. Fibros. 2015.
- Liang, F.; Shang, H.; Jordan, N.J.; Wong, E.; Mercadante, D.; Saltz, J.; Mahiou, J.; Bihler, H.J.; Mense, M. High-Throughput Screening for Readthrough Modulators of CFTR PTC Mutations. SLAS Technol. 2017.
- Giuliano, K.A.; Wachi, S.; Drew, L.; Dukovski, D.; Green, O.; Bastos, C.; Cullen, M.D.; Hauck, S.; Tait, B.D.; Munoz, B.; et al. Use of a High-Throughput Phenotypic Screening Strategy to Identify Amplifiers, a Novel Pharmacological Class of Small Molecules That Exhibit Functional Synergy with Potentiators and Correctors. SLAS Discov. 2018.
- Van Der Plas, S.E.; Kelgtermans, H.; De Munck, T.; Martina, S.L.X.; Dropsit, S.; Quinton, E.; De Blieck, A.; Joannesse, C.; Tomaskovic, L.; Jans, M.; et al. Discovery of N-(3-Carbamoyl-5,5,7,7-tetramethyl-5,7-dihydro-4H-thienopyran-2-yl)-lH-pyrazole-5-carboxamide (GLPG1837), a Novel Potentiator Which Can Open Class III Mutant Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Channels to a High Extent. J. Med. Chem. 2018, 61, 1425–1435.
- Veit, G.; Xu, H.; Dreano, E.; Avramescu, R.G.; Bagdany, M.; Beitel, L.K.; Roldan, A.; Hancock, M.A.; Lay, C.; Li, W.; et al. Structure-guided combination therapy to potently improve the function of mutant CFTRs. Nat. Med. 2018.
- Wang, X.; Liu, B.; Searle, X.; Yeung, C.; Bogdan, A.; Greszler, S.; Singh, A.; Fan, Y.; Swensen, A.M.; Vortherms, T.; et al. Discovery of 4-carbonyl}amino)-7-(difluoromethoxy)-3,4-dihydro-2H-chromen-2-yl]benzoic Acid (ABBV/GLPG-2222), a Potent Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Corrector for the Treatment of Cystic Fibrosis. J. Med. Chem. 2018, 61, 1436–1449.
- Wang, X.; Venable, J.; LaPointe, P.; Hutt, D.M.; Koulov, A.V.; Coppinger, J.; Gurkan, C.; Kellner, W.; Matteson, J.; Plutner, H.; et al. Hsp90 Cochaperone Aha1 Downregulation Rescues Misfolding of CFTR in Cystic Fibrosis. Cell 2006.
- Pankow, S.; Bamberger, C.; Calzolari, D.; Martínez-Bartolomé, S.; Lavallée-Adam, M.; Balch, W.E.; Yates, J.R. Δf508 CFTR interactome remodelling promotes rescue of cystic fibrosis. Nature 2015, 528, 510–516.
- Santos, J.D.; Canato, S.; Carvalho, A.S.; Botelho, H.M.; Aloria, K.; Amaral, M.D.; Matthiesen, R.; Falcao, A.O.; Farinha, C.M. Folding Status Is Determinant over Traffic-Competence in Defining CFTR Interactors in the Endoplasmic Reticulum. Cells 2019, 8, 353.
- Matos, A.M.; Pinto, F.R.; Barros, P.; Amaral, M.D.; Pepperkok, R.; Matos, P. Inhibition of calpain 1 restores plasma membrane stability to pharmacologically rescued Phe508del-CFTR variant. J. Biol. Chem. 2019, 294, 13396–13410.
- Novère, N.L.; Hucka, M.; Mi, H.; Moodie, S.; Schreiber, F.; Sorokin, A.; Demir, E.; Wegner, K.; Aladjem, M.I.; Wimalaratne, S.M.; et al. The Systems Biology Graphical Notation. Nat. Biotechnol. 2009, 27, 735–741.
