Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Cell Biology
Lymphatic endothelial cells (LECs) line the lymphatic vasculature and play a central role in the immune response. LECs have abilities to regulate immune transport, to promote immune cell survival, and to cross present antigens to dendritic cells. Single-cell RNA sequencing (scRNA) technology has accelerated new discoveries in the field of lymphatic vascular biology.
- lymphatics
- lymphatic endothelial cells
- single cell RNA-sequencing
1. Introduction
The lymphatic vasculature plays an essential role in many physiological and pathological processes in the human body. Although the origin of the lymphatic vasculature is closely connected to the vascular system, it has unique tasks and properties. Amongst these are the regulation of interstitial fluid balance, clearance of tissue-derived debris, and lipids and immune cell transport. Thus far, tissue fluid homeostasis has been the best studied function of the lymphatic vasculature, but novel roles are being increasingly discovered [1][2][3]. Tissue inflammation or injury, featured by increased interstitial fluid and enhanced influx of immune cells, requires expansion of the lymphatic vasculature to facilitate transport of tissue debris, leukocytes, and antigens (Ag), either soluble or carried by Ag-presenting cells, to the lymph node (LN). These events lead to an efficient immune response, and, ultimately, resolution of inflammation. In the last 20 years, identification of lymphatic markers, new experimental in vivo models, and 2D and 3D culture assays with lymphatic endothelial cells (LECs) have advanced our knowledge on lymphatic vascular biology [4]. This has important implications for a wide spectrum of disease areas, such as wound healing, obesity, cancer, and chronic inflammation and autoimmunity [5][6][7][8]. Changes in lymphatic vasculature, such as dilated lymphatic vessels or decreased amount of lymphatic vessels, are also widely observed in chronic (rheumatic) inflammatory diseases, such as psoriasis, rheumatoid arthritis (RA), systemic sclerosis, and inflammatory bowel disease (IBD) [9][10][11][12].
The recently developed technique of single-cell RNA sequencing (scRNA-seq) is widely used to uncover the identity and heterogeneity of cell subpopulations, and it has, either alone or combined with other state-of-the-art techniques, increased our knowledge on new functions of LECs in health and disease. The scRNA-seq technique evolved from sequencing only a handful of cells to currently sequencing more than 100,000 cells per sample [13]. ScRNA-seq protocols usually consist of the following steps: single cell capture, single cell lysis, reverse transcription, preamplification, library preparation, and sequencing. Laser capture microdissection, fluorescence-activated cell sorting (FACS) in microtiter plates, microfluidics, and microdroplets can be used to capture single cells. Techniques can differ in the number of cells that can be included, sequencing depth, and costs [14]. High dimensional data analysis has been applied to obtain biologically meaningful results. Frequently used strategies include dimensionality reduction methods, such as principal component analysis (PCA), t-distributed stochastic neighbor embedding (t-SNE), and uniform manifold approximation and projection (UMAP), followed by clustering algorithms, such as hierarchical clustering or k-means clustering [15]. Other promising approaches, such as trajectory inference and cell–cell interaction plots, intend to predict the origin of cells and interactions with other cells [16][17].
2. Single-Cell Transcriptomic Insides in Embryonic Development
Studies on embryonic development can often explain complex pathological processes, as has been demonstrated by Burchill et al. using scRNA-seq of liver LECs in an inflammatory environment [18].
2.1. Peripheral Lymphatic Vessel Formation
In mammals, both the origin of LECs lining the lymphatic vessels and the underlying molecular mechanisms and proteins involved in the embryonic development are not entirely clear. Historically, LECs were thought to exclusively originate from embryonic venous cells [19][20][21][22]. However, more recent studies also found that specific lymphatic vascular beds in the skin, the gut, and the heart can bear LECs from non-venous origins [23][24][25][26]. Molecular heterogeneity between LECs isolated from different organs also supports the possibility of multiple sources for LECs [27][28][29]. Unfortunately, the scRNA-seq technique has not yet been used to study the origin of LECs in the different lymphatic vascular beds.
The underlying molecular mechanisms and contributors of the venous-to-lymphatic transition have been best studied. This process starts with the transcription factor Nuclear Receptor Subfamily2 Group F Member 2 (NR2F2), also called COUP-TFII, cooperating with SRY-Box Transcription factor 18 (SOX18) to activate Prospero homeobox protein 1 (PROX1) expression [30]. PROX1 is the master transcription factor in LECs and drives expression of key markers, such as vascular endothelial growth factor receptor 3 (VEGFR-3), integrin-9alpha (adhesion molecule), podoplanin (cell migration), and chemokine (C-C motif) ligand 21 (CCL21) (chemokine) [20][31][32][33]. Downregulation of Prox1 and its target genes result in loss of LEC identity and function, as was observed in a scRNA-seq mouse liver study [18]. According to this study, LEC identity and function could be regained by exposing them to VEGF-C. VEGF-C is one of the five members of the VEGF family of angiogenic cytokines that signals through VEGFR-3, which, in turn, signals via the growth factor-regulated phosphoinositide 3-kinase (PI3K)–AKT signaling network or the Mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinases (ERK) pathways to regulate embryonic development of LECs and LEC identity, survival, proliferation, and migration in adulthood [34].
Based on data of scRNA-seq studies, new genes have been suggested to play a role in embryonic lymphatic development. A recent scRNA-seq study in a murine newborn lung model predicted, aside from Prox-1, Sox18, and Nr2f2, for example, homeobox d 8 (Hoxd8), Maf, T-box 1 (Tbx1), and ETS Transcription Factor 3 (Elk3) to be involved in LEC development [35]. Hoxd8 is a homeodomain transcription factor and can induce and maintain Prox-1 expression [30]. Maf is a transcription factor upregulated by VEGF-C/VEGFR3 signaling that plays a role in developmental lymphangiogenesis [36]. Tbx1 belongs to the T-box transcription factors, which are mostly involved in developmental processes, and Tbx1 mouse mutants have severe problems with pharyngeal and cardiovascular development. Interestingly, conditional deletion of Tbx1 in mice also causes widespread lymphangiogenesis defects due to its role in regulating VEGFR3 expression [37]. Elk3 is an Erythroblast Transformation Specific (ETS) binding domain transcription factor and has not yet been studied in the context of embryonic development. However, ELK3-expressing LECs promote breast cancer growth and migration of cancer cells [38]. As published earlier, NR2F2 can form heterodimers with PROX-1 to activate Notch signaling in LECs important for lymphangiogenesis [39]. Other unknown protein-coding genes found in the scRNA-seq in the newborn lung study include, for example, Profilin 1 (Pfn1) and AE Binding Protein 1(Aebp1) [35]. Future studies into these genes might confirm roles for these factors in embryonic development.
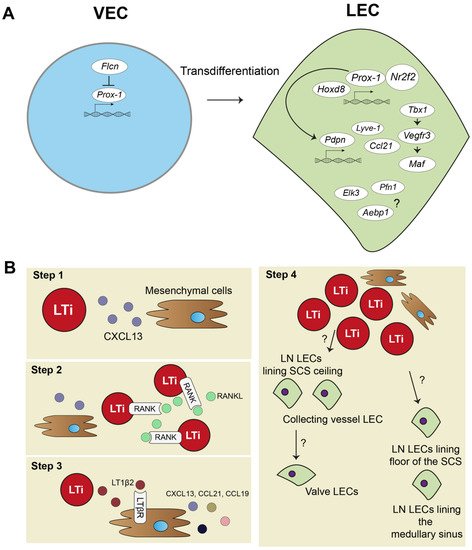
Strikingly, a recent study combining an scRNA-seq approach validated by experimental mouse models discovered that the protein Folliculin (FLCN), long known as a tumor suppressor protein, plays a major role in controlling venous-to-lymphatic cell transition. FLCN prevents expression of Prox-1 in venous endothelial cells (VECs) by binding of the Prox-1 regulatory element, the transcription factor E3 (Tfe3). FLCN deficiency results in disrupted lymphatic and venous development and dilatation of vessels [40]. Figure 1A shows an overview of venous-to-lymphatic transdifferentiation. Many processes in embryonic development driven by these transcription factors have been implicated in post-natal lymphangiogenesis during inflammation. However, knowledge on aberrant expression of many of these transcription factors is lacking.

Figure 1. Lymphatic endothelial cell and lymph node embryonic development. (A) In embryonic development, lymphatic endothelial cells (LECs) mostly transdifferentiate from venous endothelial cells (VECs). Prospero homeobox protein 1 (Prox-1) is a master regulator of lymphatic fate. Homeobox d 8 (Hoxd8) can activate Prox-1 expression, and Prox-1 cooperates with Nuclear Receptor Subfamily2 Group F Member 2 (Nr2f2) to drive expression of other key peripheral LEC molecules, such as podoplanin(Pdpn), lymphatic vessel endothelial hyaluron receptor 1 (Lyve-1), chemokine (C-C motif) ligand 21 (Ccl21), and vascular endothelial growth factor receptor 3 (Vegfr3). Vegfr3 is also regulated by transcription factor T-box 1 (Tbx1) and can activate other transcription factors, such as Maf. Single cell RNA sequencing (scRNA-seq) data also suggested transcription factors, such as ETS Transcription Factor 3 (Elk3), or other protein coding RNAs, such as Profilin 1 (Pfn1) and AE binding protein 1 (Aebp1), to be involved in lymphatic development, but their function is not yet clear. (B) Lymph node (LN) development follows several steps: (1) mesenchymal cells secrete chemokine (C-X-C motif) ligand 13 (CXCL13) to attract lymphoid tissue inducer (LTi) cells. (2) LTi cells cluster together and activate receptor activator nuclear factor κ B (RANK) and RANK ligand (RANKL) signaling. (3) RANK-RANKL signaling increases lymphotoxin α1β2 (LT1β2), which activates lymphotoxin beta receptor (LTβR) on mesenchymal cells responding with more cytokine signaling and attracting more LTi cells. (4) Mesenchymal cells differentiate into stromal cell subsets of the LN, and LTi differentiate or attract LECs. Trajectory inference of scRNA-seq data showed that there are two developmental paths of the LN LECs.
2.2. Lymph Node Development
The embryonic development of the lymph node (LN) is a step-by-step process and requires close interaction between mesenchymal cells and lymphoid tissue inducer (LTi) cells (Figure 1B). First, mesenchymal cells recruit LTi cells by expressing chemokine (C-X-C motif) ligand 13 (CXCL13) (Figure 1B, step 1). LTi cells cluster together and are identified by expression of chemokine (C-X-C motif) receptor 5 (CXCR5), the receptor for CXCL13, CD45, CD4, and CD127 (Figure 1B, step 2). LTi cells also express receptor activator nuclear factor κ B (RANK) and its ligand, RANKL, which induces high amounts of lymphotoxin α1β2 (LT1β2) [41][42][43]. LT1β2 activates the lymphtoxin beta receptor (LTβR) on mesenchymal cells. Subsequently, LTβR induces expression of cell adhesion molecules, such as intracellular adhesion molecule 1 (ICAM-1), and chemokines, such as CXCL13, CCL21, and CCL19, by mesenchymal cells. This initiates a positive feedback loop that attracts more LTi cells to the site (Figure 1B, step 3). The mesenchymal cells differentiate eventually in the stromal cells found in the lymph node. Whether LTi cells differentiate into LECs or, at their turn, attract LECs to the LN site is currently unknown [44]. Developed LNs consist of a subcapular sinus (SCS), which is connected to the afferent lymphatic vessels, and they collect migrating immune cells from draining tissues, including T- and B-lymphocytes and dendritic cells (DCs) [8]. Resident CD169+ macrophages and DCs in the SCS of the LN provide Ags, assembled from captured pathogens and molecules/proteins, to passing lymphocytes [45][46][47]. This is an essential interaction for educating lymphocytes towards proper immune responses. Lymphocytes are activated in LN parenchyma and travel through the cortical sinus (CS) and medullary sinus (MS) before they exit the LN into the peripheral blood [48].
Trajectory inference analysis of lymph node LECs predicted that LECs from the ceiling of the SCS and LECs from collecting lymphatic vessels have a shared origin, and they give rise to lymphatic valve cells (Figure 1B, step 4). LECs lining the floor of the SCS and in the MS also seem to have the same origin. No functional experiments have been performed yet to confirm these predicted developmental paths of LN LECs. Better insights in LN development are needed and will also help to further determine how tertiary lymphoid organs (TLOs) develop. TLOs, induced by LTi cells, are frequently found in chronically inflamed tissue, where they regulate inflammation and consist of many stromal cells, also found in lymph nodes [49]. Thus far, TLOs have not been investigated with scRNA-seq methodology yet.
3. The Heterogenous Family of LECs
3.1. Lymph Node LEC Subpopulations
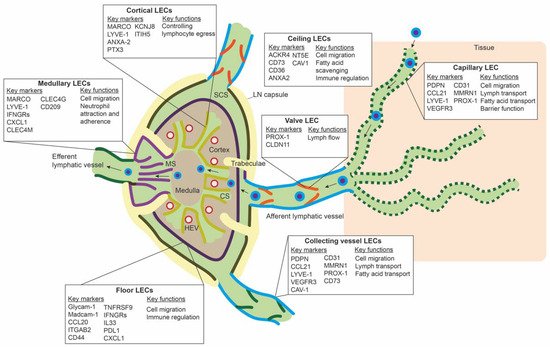
LECs mark the outline of SCS, CS, and MS in the LN, along with other stromal cells, such as fibroblastic reticular cells and perivascular cells [50]. ScRNA-seq studies have revealed transcriptomic differences between LEC populations lining these different parts of the LN, and they have identified subpopulations that delineate the floor of SCS (floor LECs), the ceiling of the SCS (ceiling LECs), the medullary sinus (medullary LECs), and the cortical sinus (cortical LECs) [51][52][53]. Figure 2 gives an overview of the different subpopulations, their spatial location, markers, and proposed functions. Differences between mice and human LN subpopulations may be species-specific, but they are more likely the result of technical differences related to the scRNA-seq technique or sequencing depth.

Figure 2. Overview of the different lymph node lymphatic endothelial cells and peripheral lymphatic endothelial cell subpopulations. The lymph node (LN) consists of multiple sinuses to transport lymph and immune cells from the afferent lymphatic vessels to the efferent lymphatic vessel. Immune cells arrive in the subcapular sinus (SCS) and migrate over the floor of the SCS, lined by floor LECs, to the LN cortex. The SCS is a narrow sinus, and it is close to the LN capsule. The LN capsule side of the SCS is lined by ceiling LECs. In the LN cortex, immune cells also arrive via high endothelial venules (HEVs). Egress of immune cells from the cortex is via the cortical sinuses (CS) lined by cortical LECs. These cortical sinuses drain in the medulla. Medullary sinuses (MS), lined by medullary LECs, transport lymph and immune cells to the efferent lymphatic vessel. LECs lining the different sinuses were found to be transcriptionally different and to have a range of different functions.
3.2. Peripheral LEC Subpopulations
In the periphery, lymphatic capillaries resorb tissue fluid, antigens, and growth factors and enable transport of migration immune cells by means of their oak-leaf shape and button-like junctions and a discontinuous basal membrane by which leukocytes or macromolecules can pass (Figure 2) [47][54][55]. Smooth muscle cells control further propulsion of lymph to the collecting lymphatic vessels. Intraluminal valves ensure unidirectional flow of lymph in the collecting vessels [56].
ScRNA-seq data of peripheral LECs is limited. Takeda et al. sequenced a few LN surrounding collecting lymphatic vessels and lymphatic capillaries. The LECs from the afferent collecting vessels shared the expression of CD73 and CAV1 with ceiling LECs of the SCS. PDPN, LYVE-1, and CCL21 were highly expressed on lymphatic capillaries, and CLDN11 was found as a marker for valve LECs [53]. It is unclear if LEC subpopulations within capillaries or collecting vessels exist. Furthermore, if LECs populations differ between organs is unknown. However, in a large scRNA-seq study of endothelial cells in mouse organs, LECs from different organs clustered together and did not seem transcriptionally different [57]. Another large mouse endothelial scRNA-seq study also observed minimal differences between LECs from different tissues, except for LECs from the gut [58].
Multiple scRNA-seq studies sequencing whole organs or tissues have found an LEC population identified by the expression of LYVE-1, PROX-1, PDPN, VEGFR3, and CCL21 [58][59][60][61][62]. In these studies, a few additional molecules were found to be expressed by LECs. Murine LECs in multiple organs express other molecules, such as Thy1, Mmrn1, and Esam, mostly involved in extra cellular matrix remodeling and cell migration [58]. In the cornea, LECs were found to additionally express many major histocompatibility complex (MHC) I genes and angiogenic privilege factors [59]. Lung LECs highly express SEMD3D, a semaphorin that is known to guide entry of DCs into lymphatics [61][63]. In the mouse liver, LECs express Mmrn1 but also Tbx1, a molecule involved in lymphangiogenesis, and Ahnak2 [37]. Ahnak2 is a large protein, and it has been found in endothelial cells in blood–tissue barriers [64]. The LECs in the mouse liver also highly expressed Cd36, indicating their role in LDL transport [18]. LECs were also found in a scRNA-seq study of human placentas. These LECs were specifically located in the maternal portion of the placenta, the decidua basalis. Pathways for cell migration and adhesion were highly enriched in these cells, indicating a role in regulating immune cell inflex in the decidua basalis [65]. The LEC clusters in most studies only consist of at most 1% of the sequenced cells. More in-depth scRNA-seq studies will hopefully provide more insights into LECs from different organs. Functional studies into some of the found expressed markers are also lacking and could potentially uncover new functions of LECs.
This entry is adapted from the peer-reviewed paper 10.3390/ijms222111976
References
- Wiig, H.; Swartz, M.A. Interstitial Fluid and Lymph Formation and Transport: Physiological Regulation and Roles in Inflammation and Cancer. Physiol. Rev. 2012, 92, 1005–1060.
- Petrova, T.V.; Koh, G.Y. Biological functions of lymphatic vessels. Science 2020, 369, eaax4063.
- Oliver, G.; Kipnis, J.; Randolph, G.J.; Harvey, N.L. The Lymphatic Vasculature in the 21st Century: Novel Functional Roles in Homeostasis and Disease. Cell 2020, 182, 270–296.
- Tewalt, E.F.; Cohen, J.N.; Rouhani, S.J.; Engelhard, V.H. Lymphatic endothelial cells-key players in regulation of tolerance and immunity. Front. Immunol. 2012, 3, 305.
- Förster, R.; Braun, A.; Worbs, T. Lymph node homing of T cells and dendritic cells via afferent lymphatics. Trends Immunol. 2012, 33, 271–280.
- Randolph, G.J.; Ivanov, S.; Zinselmeyer, B.; Scallan, J.P. The Lymphatic System: Integral Roles in Immunity. Annu. Rev. Immunol. 2017, 35, 31–52.
- Worbs, T.; Hammerschmidt, S.I.; Förster, R. Dendritic cell migration in health and disease. Nat. Rev. Immunol. 2016, 17, 30–48.
- Yeo, K.P.; Angeli, V. Bidirectional Crosstalk between Lymphatic Endothelial Cell and T Cell and Its Implications in Tumor Immunity. Front. Immunol. 2017, 8, 83.
- Kunstfeld, R.; Hirakawa, S.; Hong, Y.-K.; Schacht, V.; Lange-Asschenfeldt, B.; Velasco, P.; Lin, C.; Fiebiger, E.; Wei, X.; Wu, Y.; et al. Induction of cutaneous delayed-type hypersensitivity reactions in VEGF-A transgenic mice results in chronic skin inflammation associated with persistent lymphatic hyperplasia. Blood 2004, 104, 1048–1057.
- Bouta, E.M.; Bell, R.D.; Rahimi, H.; Xing, L.; Wood, R.W.; Iii, C.O.B.; Ritchlin, C.; Schwarz, E.M. Targeting lymphatic function as a novel therapeutic intervention for rheumatoid arthritis. Nat. Rev. Rheumatol. 2018, 14, 94–106.
- Manetti, M.; Milia, A.F.; Guiducci, S.; Romano, E.; Matucci-Cerinic, M.; Ibba-Manneschi, L. Progressive Loss of Lymphatic Vessels in Skin of Patients with Systemic Sclerosis. J. Rheumatol. 2010, 38, 297–301.
- Randolph, G.J.; Bala, S.; Rahier, J.-F.; Johnson, M.W.; Wang, P.L.; Nalbantoglu, I.; Dubuquoy, L.; Chau, A.; Pariente, B.; Kartheuser, A.; et al. Lymphoid Aggregates Remodel Lymphatic Collecting Vessels that Serve Mesenteric Lymph Nodes in Crohn Disease. Am. J. Pathol. 2016, 186, 3066–3073.
- Svensson, V.; Vento-Tormo, R.; Teichmann, S.A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 2018, 13, 599–604.
- Kolodziejczyk, A.; Kim, J.K.; Svensson, V.; Marioni, J.; Teichmann, S.A. The Technology and Biology of Single-Cell RNA Sequencing. Mol. Cell 2015, 58, 610–620.
- Luecken, M.D.; Theis, F.J. Current best practices in single-cell RNA-seq analysis: A tutorial. Mol. Syst. Biol. 2019, 15, e8746.
- Saelens, W.; Cannoodt, R.; Todorov, H.; Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 2019, 37, 547–554.
- Armingol, E.; Officer, A.; Harismendy, O.; Lewis, N.E. Deciphering cell–cell interactions and communication from gene expression. Nat. Rev. Genet. 2021, 22, 71–88.
- Tamburini, B.A.J.; Finlon, J.M.; Gillen, A.E.; Kriss, M.S.; Riemondy, K.A.; Fu, R.; Schuyler, R.P.; Hesselberth, J.R.; Rosen, H.R.; Burchill, M.A. Chronic Liver Disease in Humans Causes Expansion and Differentiation of Liver Lymphatic Endothelial Cells. Front. Immunol. 2019, 10, 1036.
- Karkkainen, M.J.; Haiko, P.; Sainio, K.; Partanen, J.; Taipale, J.; Petrova, T.V.; Jeltsch, M.; Jackson, D.G.; Talikka, M.; Rauvala, H.; et al. Vascular endothelial growth factor C is required for sprouting of the first lymphatic vessels from embryonic veins. Nat. Immunol. 2003, 5, 74–80.
- Oliver, G. Lymphatic vasculature development. Nat. Rev. Immunol. 2004, 4, 35–45.
- Srinivasan, R.S.; Dillard, M.E.; Lagutin, O.V.; Lin, F.-J.; Tsai, S.; Tsai, M.-J.; Samokhvalov, I.M.; Oliver, G. Lineage tracing demonstrates the venous origin of the mammalian lymphatic vasculature. Genes Dev. 2007, 21, 2422–2432.
- Alitalo, K.; Tammela, T.; Petrova, T.V. Lymphangiogenesis in development and human disease. Nat. Cell Biol. 2005, 438, 946–953.
- Martinez-Corral, I.; Ulvmar, M.H.; Stanczuk, L.; Tatin, F.; Kizhatil, K.; John, S.W.; Alitalo, K.; Ortega, S.; Makinen, T. Nonvenous Origin of Dermal Lymphatic Vasculature. Circ. Res. 2015, 116, 1649–1654.
- Bower, N.I.; Koltowska, K.; Pichol-Thievend, C.; Virshup, I.; Paterson, S.; Lagendijk, A.K.; Wang, W.; Lindsey, B.W.; Bent, S.; Baek, S.; et al. Mural lymphatic endothelial cells regulate meningeal angiogenesis in the zebrafish. Nat. Neurosci. 2017, 20, 774–783.
- Stanczuk, L.; Martinez-Corral, I.; Ulvmar, M.H.; Zhang, Y.; Laviña, B.; Fruttiger, M.; Adams, R.H.; Saur, D.; Betsholtz, C.; Ortega, S.; et al. cKit Lineage Hemogenic Endothelium-Derived Cells Contribute to Mesenteric Lymphatic Vessels. Cell Rep. 2015, 10, 1708–1721.
- Klotz, L.; Norman, S.; Vieira, J.; Masters, M.; Rohling, M.; Dubé, K.N.; Bollini, S.; Matsuzaki, F.; Carr, C.A.; Riley, P.R. Cardiac lymphatics are heterogeneous in origin and respond to injury. Nat. Cell Biol. 2015, 522, 62–67.
- Garrafa, E.; Trainini, L.; Benetti, A.; Saba, E.; Fezzardi, L.; Lorusso, B.; Borghetti, P.; Bottio, T.; Ceri, E.; Portolani, N.; et al. Isolation, purification, and heterogeneity of human lymphatic endothelial cells from different tissues. Lymphology 2005, 38, 159–166.
- Mouta-Bellum, C.; Kirov, A.; Miceli-Libby, L.; Mancini, M.L.; Petrova, T.V.; Liaw, L.; Prudovsky, I.; Thorpe, P.E.; Miura, N.; Cantley, L.; et al. Organ-specific lymphangiectasia, arrested lymphatic sprouting, and maturation defects resulting from gene-targeting of the PI3K regulatory isoforms p85α, p55α, and p50α. Dev. Dyn. 2009, 238, 2670–2679.
- Norrmén, C.; Ivanov, K.I.; Cheng, J.; Zangger, N.; Delorenzi, M.; Jaquet, M.; Miura, N.; Puolakkainen, P.; Horsley, V.; Hu, J.; et al. FOXC2 controls formation and maturation of lymphatic collecting vessels through cooperation with NFATc1. J. Cell Biol. 2009, 185, 439–457.
- Harada, K.; Yamazaki, T.; Iwata, C.; Yoshimatsu, Y.; Sase, H.; Mishima, K.; Morishita, Y.; Hirashima, M.; Oike, Y.; Suda, T.; et al. Identification of targets of Prox1 during in vitro vascular differentiation from embryonic stem cells: Functional roles of HoxD8 in lymphangiogenesis. J. Cell Sci. 2009, 122, 3923–3930.
- Hong, Y.-K.; Harvey, N.; Noh, Y.-H.; Schacht, V.; Hirakawa, S.; Detmar, M.; Oliver, G. Prox1 is a master control gene in the program specifying lymphatic endothelial cell fate. Dev. Dyn. 2002, 225, 351–357.
- Wigle, J.; Oliver, G. Prox1 Function Is Required for the Development of the Murine Lymphatic System. Cell 1999, 98, 769–778.
- Wong, B.; Zecchin, A.; García-Caballero, M.; Carmeliet, P. Emerging Concepts in Organ-Specific Lymphatic Vessels and Metabolic Regulation of Lymphatic Development. Dev. Cell 2018, 45, 289–301.
- Makinen, T.; Adams, R.H.; Bailey, J.; Lu, Q.; Ziemiecki, A.; Alitalo, K.; Klein, R.; Wilkinson, G.A. PDZ interaction site in ephrinB2 is required for the remodeling of lymphatic vasculature. Genes Dev. 2005, 19, 397–410.
- Guo, M.; Du, Y.; Gokey, J.; Ray, S.; Bell, S.M.; Adam, M.; Sudha, P.; Perl, A.K.; Deshmukh, H.; Potter, S.S.; et al. Single cell RNA analysis identifies cellular heterogeneity and adaptive responses of the lung at birth. Nat. Commun. 2019, 10, 1–16.
- Dieterich, L.C.; Tacconi, C.; Menzi, F.; Proulx, S.T.; Kapaklikaya, K.; Hamada, M.; Takahashi, S.; Detmar, M. Lymphatic MAFB regulates vascular patterning during developmental and pathological lymphangiogenesis. Angiogenesis 2020, 23, 411–423.
- Chen, L.; Mupo, A.; Huynh, T.; Cioffi, S.; Woods, M.; Jin, C.; McKeehan, W.; Thompson-Snipes, L.; Baldini, A.; Illingworth, E. Tbx1 regulates Vegfr3 and is required for lymphatic vessel development. J. Cell Biol. 2010, 189, 417–424.
- Kim, K.-S.; Park, J.-H.; Oh, N.; Cho, H.-J.; Park, K.-S. ELK3 expressed in lymphatic endothelial cells promotes breast cancer progression and metastasis through exosomal miRNAs. Sci. Rep. 2019, 9, 1–10.
- Aranguren, X.L.; Beerebs, M.; Coppiello, G.; Wiese, C.; Vandersnissen, I.; Nigro, A.L.; Verfaillie, C.M.; Gessler, M.; Luttun, A. COUP-TFII orchestrates venous and lymphatic endothelial identity by homo- or hetero-dimerisation with PROX1. J. Cell Sci. 2013, 126, 1164–1175.
- Tai-Nagara, I.; Hasumi, Y.; Kusumoto, D.; Hasumi, H.; Okabe, K.; Ando, T.; Matsuzaki, F.; Itoh, F.; Saya, H.; Liu, C.; et al. Blood and lymphatic systems are segregated by the FLCN tumor suppressor. Nat. Commun. 2020, 11, 1–12.
- Drayton, D.L.; Liao, S.; Mounzer, R.H.; Ruddle, N.H. Lymphoid organ development: From ontogeny to neogenesis. Nat. Immunol. 2006, 7, 344–353.
- Mebius, R.E. Organogenesis of lymphoid tissues. Nat. Rev. Immunol. 2003, 3, 292–303.
- Vondenhoff, M.F.; Greuter, M.; Goverse, G.; Elewaut, D.; Dewint, P.; Ware, C.F.; Hoorweg, K.; Kraal, G.; Mebius, R.E. LTbetaR signaling induces cytokine expression and up-regulates lymphangiogenic factors in lymph node anlagen. J. Immunol. 2009, 182, 5439–5445.
- Cupedo, T.; Mebius, R.E. Cellular Interactions in Lymph Node Development. J. Immunol. 2004, 174, 21–25.
- Carrasco, Y.R.; Batista, F.D. B Cells Acquire Particulate Antigen in a Macrophage-Rich Area at the Boundary between the Follicle and the Subcapsular Sinus of the Lymph Node. Immunity 2007, 27, 160–171.
- Gerner, M.Y.; Torabi-Parizi, P.; Germain, R.N. Strategically Localized Dendritic Cells Promote Rapid T Cell Responses to Lymph-Borne Particulate Antigens. Immunity 2015, 42, 172–185.
- Tammela, T.; Alitalo, K. Lymphangiogenesis: Molecular Mechanisms and Future Promise. Cell 2010, 140, 460–476.
- Hampton, H.R.; Chtanova, T. Lymphatic Migration of Immune Cells. Front. Immunol. 2019, 10, 1168.
- Luo, S.; Zhu, R.; Yu, T.; Fan, H.; Hu, Y.; Mohanta, S.K.; Hu, D. Chronic Inflammation: A Common Promoter in Tertiary Lymphoid Organ Neogenesis. Front. Immunol. 2019, 10, 2938.
- Rodda, L.B.; Lu, E.; Bennett, M.; Sokol, C.L.; Wang, X.; Luther, S.; Barres, B.A.; Luster, A.D.; Ye, C.J.; Cyster, J.G. Single-Cell RNA Sequencing of Lymph Node Stromal Cells Reveals Niche-Associated Heterogeneity. Immunity 2018, 48, 1014–1028.e6.
- Sibler, E.; He, Y.; Ducoli, L.; Keller, N.; Fujimoto, N.; Dieterich, L.C.; Detmar, M. Single-cell transcriptional heterogeneity of lymphatic endothelial cells in normal and inflamed murine lymph nodes. Cells 2021, 10, 1371.
- Fujimoto, N.; He, Y.; D’Addio, M.; Tacconi, C.; Detmar, M.; Dieterich, L.C. Single-cell mapping reveals new markers and functions of lymphatic endothelial cells in lymph nodes. PLoS Biol. 2020, 18, e3000704.
- Takeda, A.; Hollmén, M.; Dermadi, D.; Pan, J.; Brulois, K.F.; Kaukonen, R.; Lönnberg, T.; Boström, P.; Koskivuo, I.; Irjala, H.; et al. Single-Cell Survey of Human Lymphatics Unveils Marked Endothelial Cell Heterogeneity and Mechanisms of Homing for Neutrophils. Immunity 2019, 51, 561–572.e5.
- Baluk, P.; Fuxe, J.; Hashizume, H.; Romano, T.; Lashnits, E.; Butz, S.; Vestweber, D.; Corada, M.; Molendini, C.; Dejana, E.; et al. Functionally specialized junctions between endothelial cells of lymphatic vessels. J. Exp. Med. 2007, 204, 2349–2362.
- Tammela, T.; Saaristo, A.; Holopainen, T.; Lyytikkä, J.; Kotronen, A.; Pitkonen, M.; Abo-Ramadan, U.; Ylä-Herttuala, S.; Petrova, T.V.; Alitalo, K. Therapeutic differentiation and maturation of lymphatic vessels after lymph node dissection and transplantation. Nat. Med. 2007, 13, 1458–1466.
- Escobedo, N.; Oliver, G. Lymphangiogenesis: Origin, Specification, and Cell Fate Determination. Annu. Rev. Cell Dev. Biol. 2016, 32, 677–691.
- Feng, W.; Chen, L.; Nguyen, P.K.; Wu, S.M.; Li, G. Single Cell Analysis of Endothelial Cells Identified Organ-Specific Molecular Signatures and Heart-Specific Cell Populations and Molecular Features. Front. Cardiovasc. Med. 2019, 6, 165.
- Kalucka, J.; de Rooij, L.P.M.H.; Goveia, J.; Rohlenova, K.; Dumas, S.J.; Meta, E.; Conchinha, N.; Taverna, F.; Teuwen, L.-A.; Veys, K.; et al. Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell 2020, 180, 764–779.e20.
- Duong, T.; Koltowska, K.; Pichol-Thievend, C.; Le Guen, L.; Fontaine, F.; Smith, K.A.; Truong, V.; Skoczylas, R.; Stacker, S.A.; Achen, M.G.; et al. VEGFD regulates blood vascular development by modulating SOX18 activity. Blood 2014, 123, 1102–1112.
- Kalluri, A.; Vellarikkal, S.K.; Edelman, E.; Nguyen, L.; Subramanian, A.; Ellinor, P.T.; Regev, A.; Kathiresan, S.; Gupta, R.M. Single-Cell Analysis of the Normal Mouse Aorta Reveals Functionally Distinct Endothelial Cell Populations. Circculation 2019, 140, 147–163.
- Schupp, J.C.; Adams, T.S.; Cosme, C.; Raredon, M.S.B.; Yuan, Y.; Omote, N.; Poli, S.; Chioccioli, M.; Rose, K.-A.; Manning, E.P.; et al. Integrated Single-Cell Atlas of Endothelial Cells of the Human Lung. Circculation 2021, 144, 286–302.
- Su, T.; Yang, Y.; Lai, S.; Jeong, J.; Jung, Y.; McConnell, M.; Utsumi, T.; Iwakiri, Y. Single-Cell Transcriptomics Reveals Zone-Specific Alterations of Liver Sinusoidal Endothelial Cells in Cirrhosis. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 1139–1161.
- Takamatsu, H.; Takegahara, N.; Nakagawa, Y.; Tomura, M.; Taniguchi, M.; Friedel, R.; Rayburn, H.; Tessier-Lavigne, M.; Yoshida, Y.; Okuno, T.; et al. Semaphorins guide the entry of dendritic cells into the lymphatics by activating myosin II. Nat. Immunol. 2010, 11, 594–600.
- Gentil, B.J.; Benaud, C.; Delphin, C.; Remy, C.; Berezowski, V.; Cecchelli, R.; Feraud, O.; Vittet, D.; Bauider, J. Specific AHNAK expression in brain endothelial cells with barrier properties. J. Cell Physiol. 2005, 203, 362–371.
- Pique-Regi, R.; Romero, R.; Tarca, A.L.; Sendler, E.D.; Xu, Y.; Garcia-Flores, V.; Leng, Y.; Luca, F.; Hassan, S.S.; Gomez-Lopez, N. Single cell transcriptional signatures of the human placenta in term and preterm parturition. eLife 2019, 8, 52004.
This entry is offline, you can click here to edit this entry!
