MicroRNAs (miRNAs) are non-coding single-stranded RNA molecules encoded by endogenous genes with ~22 nucleotides which are involved in the regulation of post-transcriptional gene expression. Ubiquitination and deubiquitination are common post-translational modifications in eukaryotic cells and important pathways in regulating protein degradation and signal transduction, in which E3 ubiquitin ligases and deubiquitinases (DUBs) play a decisive role. MiRNA and ubiquitination are involved in the regulation of most biological processes, including autophagy.
- microRNAs
- E3 ubiquitin ligases
- deubiquitinases
- autophagy
1. Introduction
2. Overview of miRNA
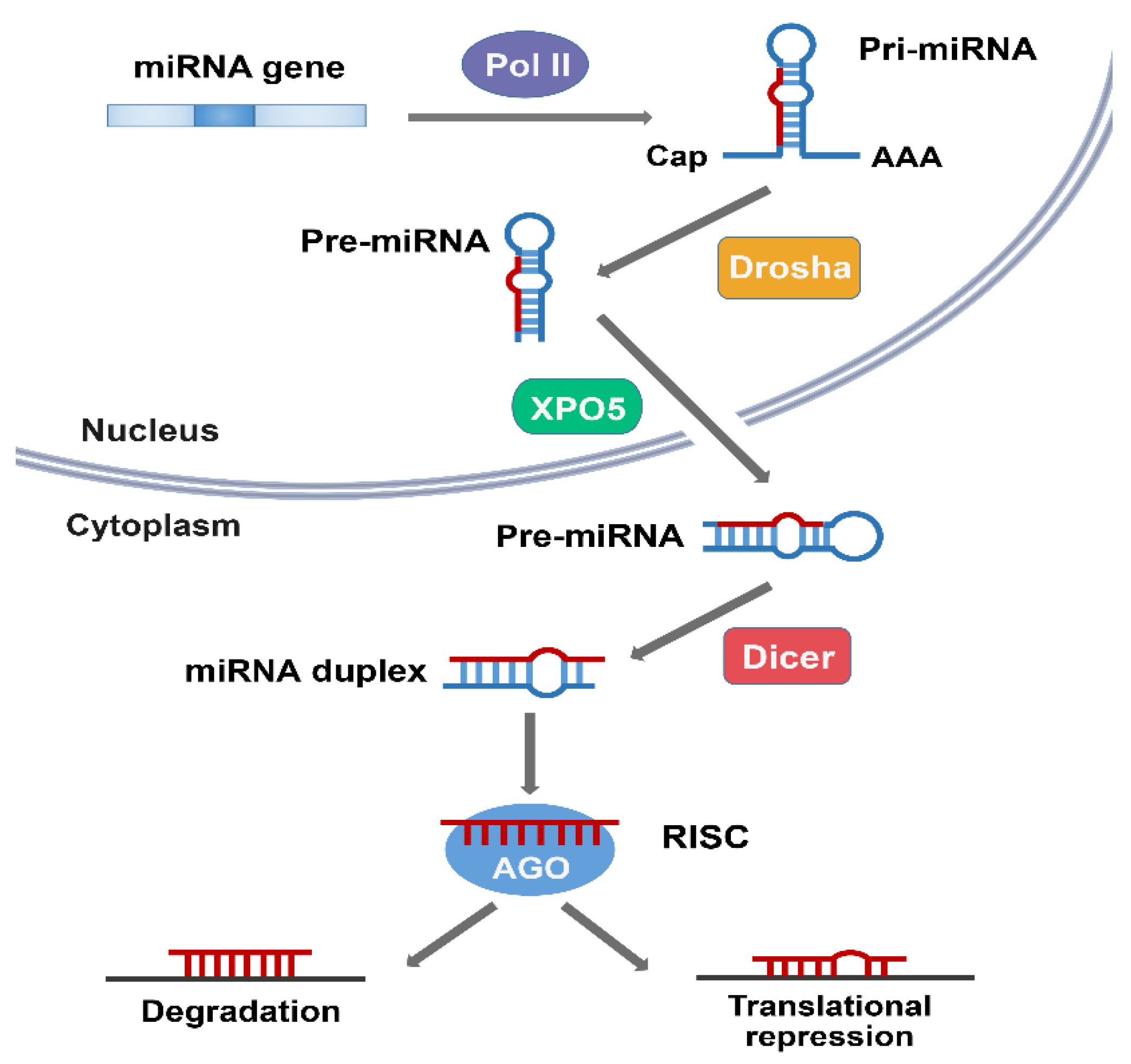
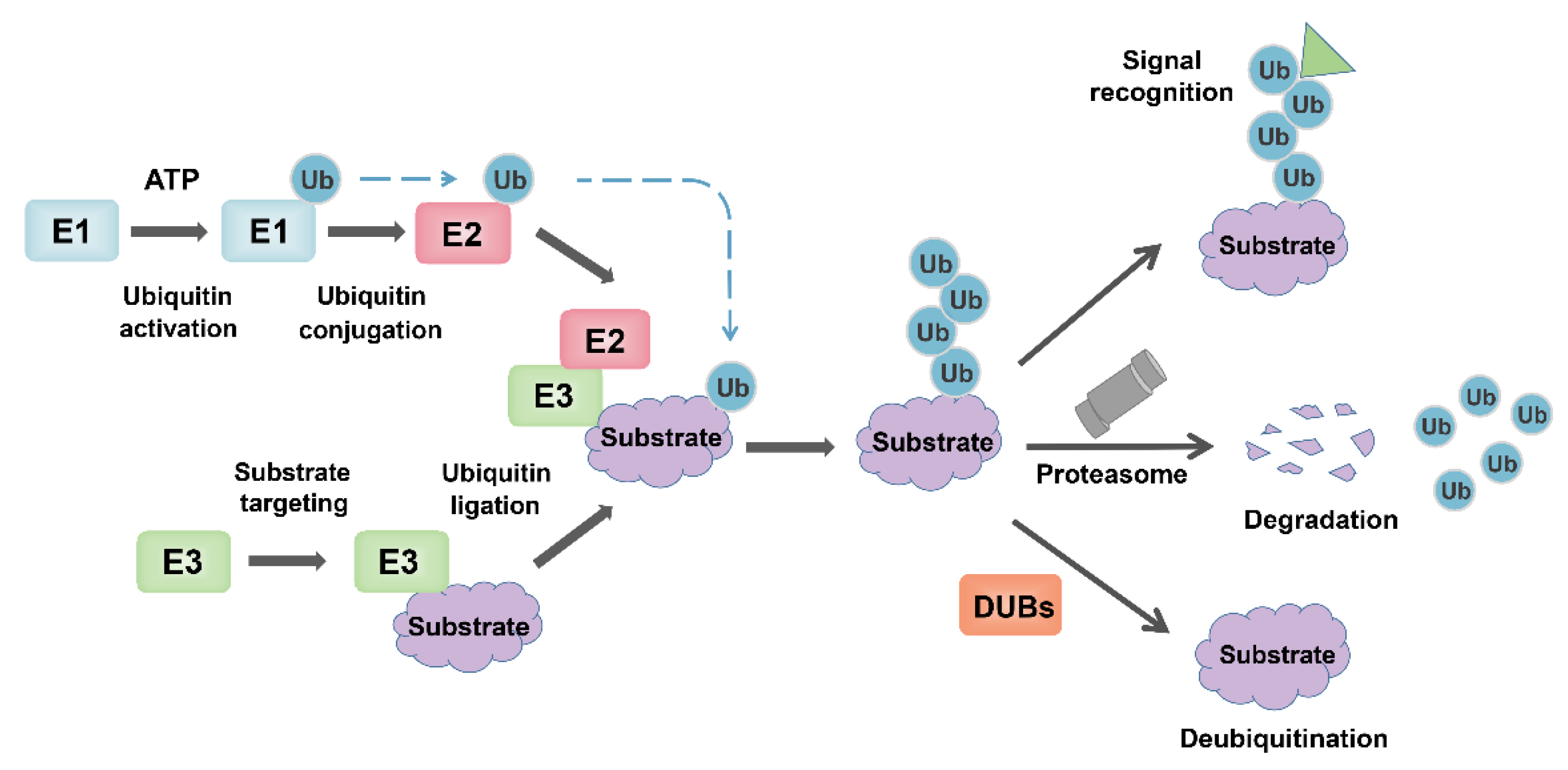
3. Overview of Ubiquitin-Proteasome System (UPS)
4. Overview of Autophagy System
5. MiRNAs Are Involved in Autophagy via Regulation of E3 Ubiquitin Ligases
|
MiRNA/E3 |
Target |
Function |
References |
|---|---|---|---|
|
Mir-30a |
MARCH5 |
MARCH5 mRNA acts as ceRNA of ATG5 |
[59] |
|
Mir-200a |
MARCH7 |
MARCH7 mRNA acts as ceRNA of ATG7 |
[60] |
|
Mir-233 |
TRIM37 |
Promotes autophagy by inhibiting MTORC1 |
[61] |
|
Mir-34a-5p |
SYVN1 |
Induces autophagy |
[62] |
|
Mir-146a |
TRAF6 |
Inhibits autophagy via ULK1 protein |
[63] |
|
Mir-27 |
NEDD4 |
Attenuates autophagy through Notch1 |
[64] |
|
TRIM65 |
Mir-138-5P |
Upregulates ATG7 by inhibiting miRISC |
[65] |
6. MiRNAs Are Involved in Autophagy via DUBs Regulation
This entry is adapted from the peer-reviewed paper 10.3390/ijms222111912
References
- Nandi, D.; Tahiliani, P.; Kumar, A.; Chandu, D. The ubiquitin-proteasome system. J. Biosci. 2006, 31, 137–155.
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422.
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513.
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157.
- Berndsen, C.E.; Wolberger, C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014, 21, 301–307.
- Ambros, V. microRNAs: Tiny regulators with great potential. Cell 2001, 107, 823–826.
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205.
- Simonson, B.; Das, S. MicroRNA Therapeutics: The Next Magic Bullet? Mini Rev. Med. Chem. 2015, 15, 467–474.
- Akkoc, Y.; Gozuacik, D. MicroRNAs as major regulators of the autophagy pathway. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118662.
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114.
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610.
- Pasquinelli, A.E.; Hunter, S.; Bracht, J. MicroRNAs: A developing story. Curr. Opin. Genet. Dev. 2005, 15, 200–205.
- Shabalina, S.A.; Spiridonov, N.A. The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol. 2004, 5, 105.
- Chekulaeva, M.; Filipowicz, W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009, 21, 452–460.
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379.
- Bentwich, I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005, 579, 5904–5910.
- Friedman, J.M.; Jones, P.A. MicroRNAs: Critical mediators of differentiation, development and disease. Swiss Med. Wkly. 2009, 139, 466–472.
- Mohr, A.M.; Mott, J.L. Overview of microRNA biology. Semin. Liver Dis. 2015, 35, 3–11.
- Han, J.; Pedersen, J.S.; Kwon, S.C.; Belair, C.D.; Kim, Y.K.; Yeom, K.H.; Yang, W.Y.; Haussler, D.; Blelloch, R.; Kim, V.N. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 2009, 136, 75–84.
- Yan, H.; Liang, F.S. miRNA inhibition by proximity-enabled Dicer inactivation. Methods 2019, 167, 117–123.
- Schwarz, D.S.; Hutvagner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208.
- Sheu-Gruttadauria, J.; MacRae, I.J. Phase Transitions in the Assembly and Function of Human miRISC. Cell 2018, 173, 946–957.e16.
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233.
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 143.
- Barwari, T.; Joshi, A.; Mayr, M. MicroRNAs in Cardiovascular Disease. J. Am. Coll. Cardiol. 2016, 68, 2577–2584.
- Davalos, V.; Esteller, M. MicroRNAs and cancer epigenetics: A macrorevolution. Curr. Opin. Oncol. 2010, 22, 35–45.
- Trang, P.; Weidhaas, J.B.; Slack, F.J. MicroRNAs as potential cancer therapeutics. Oncogene 2008, 27 (Suppl. 2), 52–57.
- Erpapazoglou, Z.; Walker, O.; Haguenauer-Tsapis, R. Versatile roles of k63-linked ubiquitin chains in trafficking. Cells 2014, 3, 1027–1088.
- Jarome, T.J.; Devulapalli, R.K. The Ubiquitin-Proteasome System and Memory: Moving Beyond Protein Degradation. Neuroscientist 2018, 24, 639–651.
- Weber, J.; Polo, S.; Maspero, E. HECT E3 Ligases: A Tale With Multiple Facets. Front. Physiol. 2019, 10, 370.
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434.
- Spratt, D.E.; Walden, H.; Shaw, G.S. RBR E3 ubiquitin ligases: New structures, new insights, new questions. Biochem. J. 2014, 458, 421–437.
- Eletr, Z.M.; Wilkinson, K.D. Regulation of proteolysis by human deubiquitinating enzymes. Biochim. Biophys. Acta 2014, 1843, 114–128.
- Magraoui, F.E.; Reidick, C.; Meyer, H.E.; Platta, H.W. Autophagy-Related Deubiquitinating Enzymes Involved in Health and Disease. Cells 2015, 4, 596–621.
- Nijman, S.M.; Luna-Vargas, M.P.; Velds, A.; Brummelkamp, T.R.; Dirac, A.M.; Sixma, T.K.; Bernards, R. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005, 123, 773–786.
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741.
- Mazure, N.M.; Pouyssegur, J. Hypoxia-induced autophagy: Cell death or cell survival? Curr. Opin. Cell Biol. 2010, 22, 177–180.
- Yu, L.; Chen, Y.; Tooze, S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018, 14, 207–215.
- Rajendran, P.; Alzahrani, A.M.; Hanieh, H.N.; Kumar, S.A.; Ben Ammar, R.; Rengarajan, T.; Alhoot, M.A. Autophagy and senescence: A new insight in selected human diseases. J. Cell. Physiol. 2019, 234, 21485–21492.
- Rubinsztein, D.C.; Marino, G.; Kroemer, G. Autophagy and aging. Cell 2011, 146, 682–695.
- Klionsky, D.J.; Codogno, P. The mechanism and physiological function of macroautophagy. J. Innate Immun. 2013, 5, 427–433.
- Killackey, S.A.; Philpott, D.J.; Girardin, S.E. Mitophagy pathways in health and disease. J. Cell Biol. 2020, 219, e202004029.
- Strappazzon, F. A global view of the miRNA-mitophagy connexion. Prog. Mol. Biol. Transl. Sci. 2020, 172, 37–54.
- Xie, Z.; Klionsky, D.J. Autophagosome formation: Core machinery and adaptations. Nat. Cell Biol. 2007, 9, 1102–1109.
- Alers, S.; Loffler, A.S.; Wesselborg, S.; Stork, B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: Cross talk, shortcuts, and feedbacks. Mol. Cell. Biol. 2012, 32, 2–11.
- Nazio, F.; Strappazzon, F.; Antonioli, M.; Bielli, P.; Cianfanelli, V.; Bordi, M.; Gretzmeier, C.; Dengjel, J.; Piacentini, M.; Fimia, G.M.; et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 2013, 15, 406–416.
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 2009, 20, 1981–1991.
- Militello, R.D.; Colombo, M.I. A membrane is born: Origin of the autophagosomal compartment. Curr. Mol. Med. 2011, 11, 197–203.
- Abrahamsen, H.; Stenmark, H.; Platta, H.W. Ubiquitination and phosphorylation of Beclin 1 and its binding partners: Tuning class III phosphatidylinositol 3-kinase activity and tumor suppression. FEBS Lett. 2012, 586, 1584–1591.
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476.
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132.
- Kotani, T.; Kirisako, H.; Koizumi, M.; Ohsumi, Y.; Nakatogawa, H. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. USA 2018, 115, 10363–10368.
- Proikas-Cezanne, T.; Takacs, Z.; Donnes, P.; Kohlbacher, O. WIPI proteins: Essential PtdIns3P effectors at the nascent autophagosome. J. Cell Sci. 2015, 128, 207–217.
- Ichimura, Y.; Kirisako, T.; Takao, T.; Satomi, Y.; Shimonishi, Y.; Ishihara, N.; Mizushima, N.; Tanida, I.; Kominami, E.; Ohsumi, M.; et al. A ubiquitin-like system mediates protein lipidation. Nature 2000, 408, 488–492.
- Rogov, V.; Dotsch, V.; Johansen, T.; Kirkin, V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 2014, 53, 167–178.
- Noda, N.N.; Satoo, K.; Fujioka, Y.; Kumeta, H.; Ogura, K.; Nakatogawa, H.; Ohsumi, Y.; Inagaki, F. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol. Cell 2011, 44, 462–475.
- Kabeya, Y.; Mizushima, N.; Yamamoto, A.; Oshitani-Okamoto, S.; Ohsumi, Y.; Yoshimori, T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004, 117, 2805–2812.
- Botti-Millet, J.; Nascimbeni, A.C.; Dupont, N.; Morel, E.; Codogno, P. Fine-tuning autophagy: From transcriptional to posttranslational regulation. Am. J. Physiol. Cell Physiol. 2016, 311, C351–C362.
- Hu, J.; Meng, Y.; Zhang, Z.; Yan, Q.; Jiang, X.; Lv, Z.; Hu, L. MARCH5 RNA promotes autophagy, migration, and invasion of ovarian cancer cells. Autophagy 2017, 13, 333–344.
- Hu, J.; Zhang, L.; Mei, Z.; Jiang, Y.; Yi, Y.; Liu, L.; Meng, Y.; Zhou, L.; Zeng, J.; Wu, H.; et al. Interaction of E3 Ubiquitin Ligase MARCH7 with Long Noncoding RNA MALAT1 and Autophagy-Related Protein ATG7 Promotes Autophagy and Invasion in Ovarian Cancer. Cell. Physiol. Biochem. 2018, 47, 654–666.
- Brigant, B.; Demont, Y.; Ouled-Haddou, H.; Metzinger-Le Meuth, V.; Testelin, S.; Garcon, L.; Metzinger, L.; Rochette, J. TRIM37 is highly expressed during mitosis in CHON-002 chondrocytes cell line and is regulated by miR-223. Bone 2020, 137, 115393.
- Tian, F.; Wang, J.; Zhang, Z.; Yang, J. LncRNA SNHG7/miR-34a-5p/SYVN1 axis plays a vital role in proliferation, apoptosis and autophagy in osteoarthritis. Biol. Res. 2020, 53, 9.
- Tang, Y.; Tao, Y.; Wang, L.; Yang, L.; Jing, Y.; Jiang, X.; Lei, L.; Yang, Z.; Wang, X.; Peng, M.; et al. NPM1 mutant maintains ULK1 protein stability via TRAF6-dependent ubiquitination to promote autophagic cell survival in leukemia. FASEB J. 2021, 35, e21192.
- Che, F.; Chen, J.; Wan, C.; Huang, X. MicroRNA-27 Inhibits Autophagy and Promotes Proliferation of Multiple Myeloma Cells by Targeting the NEDD4/Notch1 Axis. Front. Oncol. 2020, 10, 571914.
- Pan, X.; Chen, Y.; Shen, Y.; Tantai, J. Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis. 2019, 10, 429.
- Huang, L.; Hu, C.; Cao, H.; Wu, X.; Wang, R.; Lu, H.; Li, H.; Chen, H. MicroRNA-29c Increases the Chemosensitivity of Pancreatic Cancer Cells by Inhibiting USP22 Mediated Autophagy. Cell. Physiol. Biochem. 2018, 47, 747–758.
- Xiong, H.; Ni, Z.; He, J.; Jiang, S.; Li, X.; He, J.; Gong, W.; Zheng, L.; Chen, S.; Li, B.; et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene 2017, 36, 3528–3540.
- Chen, E.; Li, E.; Liu, H.; Zhou, Y.; Wen, L.; Wang, J.; Wang, Y.; Ye, L.; Liang, T. miR-26b enhances the sensitivity of hepatocellular carcinoma to Doxorubicin via USP9X-dependent degradation of p53 and regulation of autophagy. Int. J. Biol. Sci. 2021, 17, 781–795.
- Liang, H.; Su, X.; Wu, Q.; Shan, H.; Lv, L.; Yu, T.; Zhao, X.; Sun, J.; Yang, R.; Zhang, L.; et al. LncRNA 2810403D21Rik/Mirf promotes ischemic myocardial injury by regulating autophagy through targeting Mir26a. Autophagy 2020, 16, 1077–1091.
