MiRNAs are ~22-nucleotide long noncoding sequences of RNA that are located across the genome, within an intron or untranslated region (UTR) of a coding gene. Pri-miRNAs are transcribed from their genes in longer primary transcripts which are processed by two RNase III proteins—Drosha and Dicer—to form a functional miRISC complex that binds to the 3′ UTR of target mRNAs and induces their degradation and translational repression . miRNAs were found to be highly stable in blood and other body fluids, where they circulate in a cell-free form, bound to other proteins, lipids, or lipoprotein or encapsulated in exosomes. The development of specific high-throughput detection methods allowing miRNA detection in extracellular fluids, besides the fact that profiles of miRNAs were shown to be either downregulated or overexpressed across several cancer types compared to normal counterparts, has paved the way for serum miRNAs to be developed as biomarkers for early detection and monitoring of tumor evolution. However, significant challenges remain, such as the low concentration of miRNAs released in the blood, especially in early-stage disease, and the difficult identification of biomarker release sites.
- miRNAs
- biomarkers
- prostate cancer
- ultrasounds
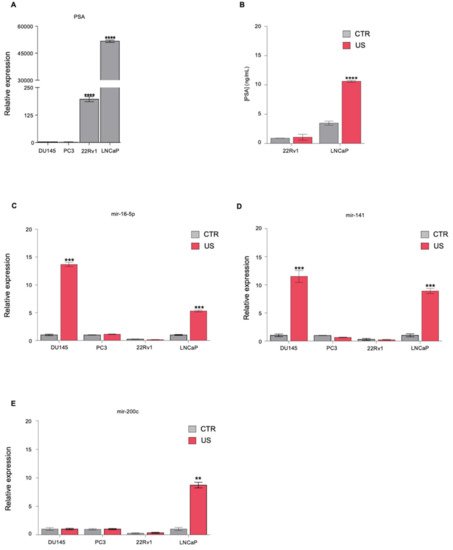
1. Ultrasound Treatment Increases the Release of Known Biomarkers in PCa Cell Lines
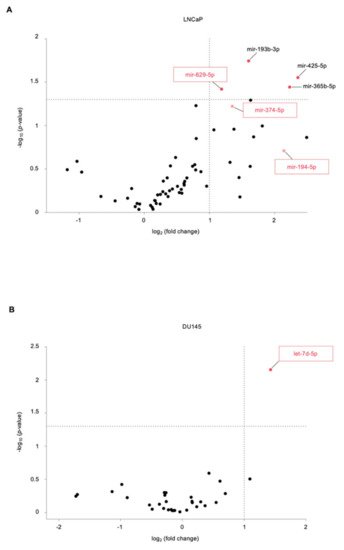
2. Identification of Novel miRNAs in the Supernatant of PCa Cells following US Treatment
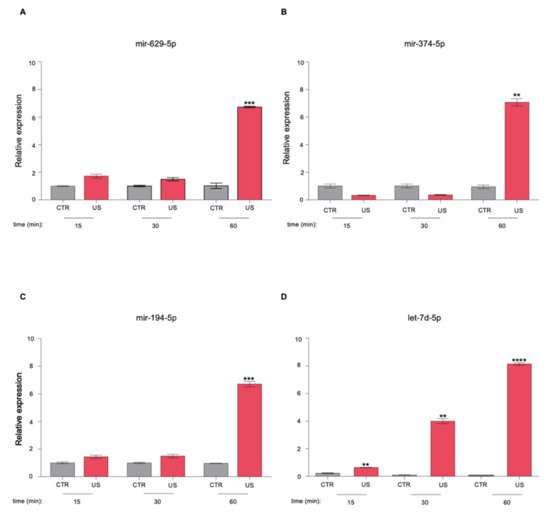
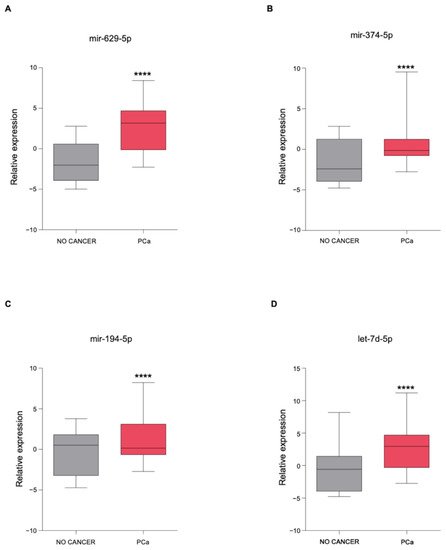
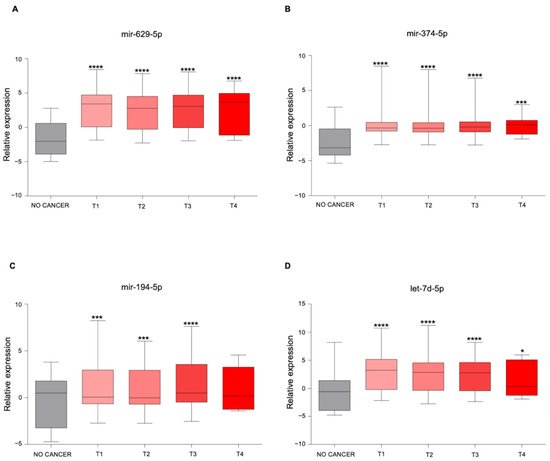
3. The Newly Identified miRNAs Are Upregulated in the Serum from PCa Patients
4. In Silico miRNA: Gene Interaction Analysis
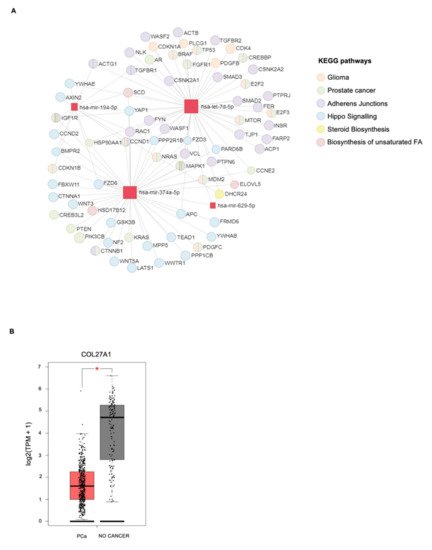
| miRNAs | KEGG Pathways | p-Value | Target Genes |
|---|---|---|---|
| miR-629-5p | Prolactin signaling pathway | 4.27 × 10−3 | PRLR AKT3 |
| miR-194-5p | Valine, leucine, and isoleucine degradation |
6.99 × 10−4 | BCKDHA ACADSB BCAT1 |
| let-7d-5p | ECM–receptor interaction | 1.07 × 10−2 | COL27A1 COL3A1 COL1A1 COL1A2 |
This entry is adapted from the peer-reviewed paper 10.3390/genes12111726
References
- Chevillet, J.R.; Khokhlova, T.D.; Giraldez, M.D.; Schade, G.R.; Starr, F.; Wang, Y.N.; Gallichotte, E.N.; Wang, K.; Hwang, J.H.; Tewari, M. Release of Cell-free MicroRNA Tumor Biomarkers into the Blood Circulation with Pulsed Focused Ultrasound: A Noninvasive, Anatomically Localized, Molecular Liquid Biopsy. Radiology 2017, 283, 158–167.
- D’Souza, A.L.; Chevillet, J.R.; Ghanouni, P.; Yan, X.; Tewari, M.; Gambhir, S.S. Tumor characterization by ultrasound-release of multiple protein and microRNA biomarkers, preclinical and clinical evidence. PLoS ONE 2018, 13, e0194268.
- Lentacker, I.; De Cock, I.; Deckers, R.; De Smedt, S.C.; Moonen, C.T.W. Understanding ultrasound induced sonoporation: Definitions and underlying mechanisms. Adv. Drug Deliv. Rev. 2014, 72, 49–64.
- Zhou, Y.; Kumon, R.E.; Cui, J.; Deng, C.X. The size of sonoporation pores on the cell membrane. Ultrasound Med. Biol. 2009, 35, 1756–1760.
- Miller, M.W.; Battaglia, L.F. The relevance of cell size on ultrasound-induced hemolysis in mouse and human blood in vitro. Ultrasound Med. Biol. 2003, 29, 1479–1485.
- Zhang, J.Y.; Su, X.P.; Li, Y.N.; Guo, Y.H. MicroRNA-425-5p promotes the development of prostate cancer via targeting forkhead box J3. Eur. Rev. Med.Pharmacol Sci. 2019, 23, 547–554.
- Lyu, J.; Zhao, L.; Wang, F.; Ji, J.; Cao, Z.; Xu, H.; Shi, X.; Zhu, Y.; Zhang, C.; Guo, F.; et al. Discovery and Validation of Serum MicroRNAs as Early Diagnostic Biomarkers for Prostate Cancer in Chinese Population. Biomed Res. Int. 2019, 2019, 9306803.
- Mazzu, Y.Z.; Yoshikawa, Y.; Nandakumar, S.; Chakraborty, G.; Armenia, J.; Jehane, L.E.; Lee, G.M.; Kantoff, P.W. Methylation-associated miR-193b silencing activates master drivers of aggressive prostate cancer. Mol. Oncol. 2019, 13, 1944–1958.
- Wagner, S.; Ngezahayo, A.; Murua Escobar, H.; Nolte, I. Role of miRNA let-7 and its major targets in prostate cancer. Biomed Res. Int. 2014, 2014, 376326.
- Battista, M.C.; Guimond, M.O.; Roberge, C.; Doueik, A.A.; Fazli, L.; Gleave, M.; Sabbagh, R.; Gallo-Payet, N. Inhibition of DHCR24/seladin-1 impairs cellular homeostasis in prostate cancer. Prostate 2010, 70, 921–933.
- Kim, S.J.; Choi, H.; Park, S.S.; Chang, C.; Kim, E. Stearoyl CoA desaturase (SCD) facilitates proliferation of prostate cancer cells through enhancement of androgen receptor transactivation. Mol. Cells 2011, 31, 371–377.
- Centenera, M.M.; Scott, J.S.; Machiels, J.; Nassar, Z.D.; Miller, D.C.; Zininos, I.; Dehairs, J.; Burvenich, I.J.G.; Zadra, G.; Chetta, P.M.; et al. ELOVL5 is a critical and targetable fatty acid elongase in prostate cancer. Cancer Res. 2021, 81, 1704–1718.
- Sun, T.; Oh, W.K.; Jacobus, S.; Regan, M.; Pomerantz, M.; Freedman, M.L.; Lee, G.S.; Kantoff, P.W. The impact of common genetic variations in genes of the sex hormone metabolic pathways on steroid hormone levels and prostate cancer aggressiveness. Cancer Prev. Res. (Phila) 2011, 4, 2044–2050.
- Chung, J.H.; Dewal, N.; Sokol, E.; Mathew, P.; Whitehead, R.; Millis, S.Z.; Frampton, G.M.; Bratslavsky, G.; Pal, S.K.; Lee, R.J.; et al. Prospective Comprehensive Genomic Profiling of Primary and Metastatic Prostate Tumors. JCO Precis Oncol. 2019, 3. PO.18.00283.
- Ikeda, S.; Elkin, S.K.; Tomson, B.N.; Carter, J.L.; Kurzrock, R. Next-generation sequencing of prostate cancer: Genomic and pathway alterations, potential actionability patterns, and relative rate of use of clinical-grade testing. Cancer Biol. Ther. 2019, 20, 219–226.
- Sowalsky, A.G.; Sager, R.; Schaefer, R.J.; Bratslavsky, G.; Pandolfi, P.P.; Balk, S.P.; Kotula, L. Loss of Wave1 gene defines a subtype of lethal prostate cancer. Oncotarget 2015, 6, 12383–12391.
- Lin, H.P.; Lin, C.Y.; Huo, C.; Jan, Y.J.; Tseng, J.C.; Jiang, S.S.; Kuo, Y.Y.; Chen, S.C.; Wang, C.T.; Chan, T.M.; et al. AKT3 promotes prostate cancer proliferation cells through regulation of Akt, B-Raf, and TSC1/TSC2. Oncotarget 2015, 6, 27097–27112.
- O’Sullivan, C.C.; Bates, S.E. Targeting Prolactin Receptor (PRLR) Signaling in PRLR-Positive Breast and Prostate Cancer. Oncologist 2016, 21, 523–526.
- Zhu, W.; Shao, Y.; Peng, Y. MicroRNA-218 inhibits tumor growth and increases chemosensitivity to CDDP treatment by targeting BCAT1 in prostate cancer. Mol. Carcinog. 2017, 56, 1570–1577.
- Pace, J.M.; Corrado, M.; Missero, C.; Byers, P.H. Identification, characterization and expression analysis of a new fibrillar collagen gene, COL27A1. Matrix Biol. 2003, 22, 3–14.
