Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biology
KCNQ1 Opposite Strand/Antisense Transcript 1 (KCNQ1OT1) encodes a lncRNA from the opposite strand of KCNQ1 in the CDKN1C/KCNQ1OT1 cluster that is reported to play a vital role in the development and progression of cancer.
- KCNQ1OT1
- human cancers
- long noncoding RNA
1. Basic Characteristics of Human Chromosome 11p15.5 and lncRNA KCNQ1OT1 Gene
KCNQ1 Opposite Strand/Antisense Transcript 1 (KCNQ1OT1), also known as KCNQ1 overlapping transcript 1, or LIT1, is a 91 kb un-spliced lncRNA located on chromosome 11p15.5 (Figure 1). The KCNQ1OT1 gene is part of a cluster of genes that undergo genomic imprinting, an epigenetic modification involving parent-specific gene expression modification. Genomic imprinting plays a critical role in fetal growth and development and is regulated by a nearby region of DNA known as imprinting center 2 (IC2) or KvDMR, which undergoes differential methylation [1][2]. The human CDKN1C/KCNQ1OT1 cluster exists as imprinted genes, expressing only one copy, with the allele activity depending on the parental origin. The paternally expressed KCNQ1OT1 transcript originates from intron 11 and is antisense to its associated protein-coding gene, Potassium Voltage-Gated Channel Subfamily Q Member 1 (KCNQ1) [3][4][5][6]. The antisense lncRNA KCNQ1OT1 promoter maps to KCNQ1 imprinting control regions, methylated on the maternal chromosome but un-methylated on the paternal chromosome.
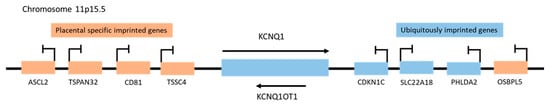
Figure 1. Schematic representation of imprinted gene clusters on human chromosome 11p15.5.
Genes located near the KCNQ1OT1 promoter (KCNQ1, CDKN1C, SLC22A18, and PHLDA2) that are imprinted both in the embryo and extra-embryonic tissues such as the placenta are ubiquitously imprinted genes, whereas the placental specific imprinted genes ASCL2, TSPAN32, CD81, TSSC4, and OSBPL5, and are only imprinted in the placenta (Figure 1) [7]. The ubiquitously expressed KCNQ1OT1 is more frequently localized in the nucleus, interacts with chromatin complexes, and regulates the genomic imprinting of multiple genes through bidirectional transcription-mediated silencing in cis [6][8][9]. Thus, the DNA sequences of the KCNQ1 and KCNQ1OT1 genes are “read” in opposite directions and have very different functions. LncRNA KCNQ1OT1 is expressed in every tissue [10] and regulates genes vital for normal growth and development before birth, as well as postnatal behavior [6][11]. However, deletion of its promoter or early transcript termination results in a loss of KCNQ1OT1 and a disruption of imprinting in the CDKN1C/KCNQ1OT1 domain, which can lead to growth-related disorders (ex. Beckwith-Wiedemann syndrome) and cancer, as well as bi-allelic expression of the entire KCNQ1 domain [12].
2. KCNQ1OT1 in Human Cancers
In the present review, we will explore current knowledge on the role of KCNQ1OT1 in the development of various human cancers. We will thoroughly discuss the molecular and mechanistic role of KCNQ1OT1 in modulating oncogenic and biological functions, regulating cancer cell signaling mechanisms, and describe how its expression correlates to clinical features (Table 1). The interaction of KCNQ1OT1 and miRNAs or/and proteins and the potential targets in different cancers are summarized in Figure 2.
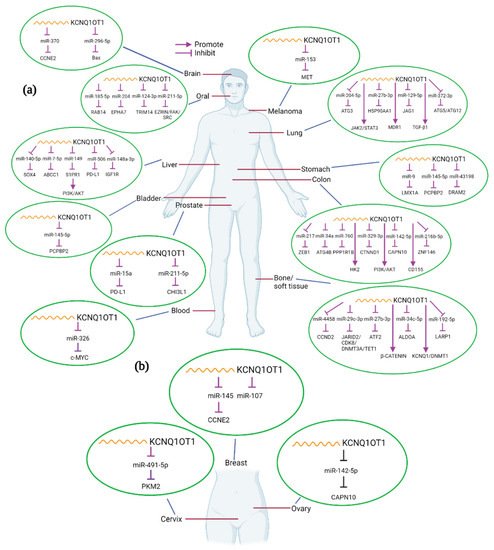
Figure 2. Interactions between KCNQ1OT1 and miRNA/genes in different kinds of human cancers. KCNQ1OT1 can interact with various miRNAs and genes in different tumor types (a), including female-specific cancers (b). Created with BioRender.com.
Table 1. Functional characterization of lncRNA KCNQ1OT1 and its targets in various cancers.
| Cancer | Expression Level | Role | Associated Clinical Features † | Functional Role † | Regulatory Molecule and Pathway [Reference] |
|---|---|---|---|---|---|
| Colorectal cancer | Upregulated | Oncogenic | Tumor size, TNM stage, lymph node metastasis, distant metastasis, histological differentiation, adjuvant therapy, primary tumor site, OS, DFS |
Proliferation, cell cycle, apoptosis, migration, invasion, aerobic glycolysis, methotrexate resistance, adjuvant fluoropyrimidine-based chemotherapy |
miR-217/ZEB1 [13]; miR-34a/Atg4B [14]; miR-760/PPP1R1B [3]; miR-329-3p/CTNND1 [15]; miR-216b-5p/ZNF146 [16]; HK2 [17]; PI3K/AKT [18]; CD155 [19] |
| Ovarian cancer | Upregulated | Oncogenic | OS | Proliferation, invasion, | miR-142-5p/CAPN10 [20] |
| Cervical cancer | Upregulated | Oncogenic | not investigated | Proliferation, metastasis, and radioresistance | miR-491-5p/PKM2 [21] |
| Glioma | Upregulated | Oncogenic | Histopathological grade | Proliferation, apoptosis, migration, invasion | miR-370/CCNE2 [22] |
| Neuroblastoma | not investigated | Suppressor | not investigated | Apoptosis | miR-296-5p/Bax [23] |
| Sarcoma | not investigated | not investigated | Histological type, metastasis, tumor depth, necrosis | not investigated | miR-29c-3p)/JARID2/CDK8/DNMT3A/TET1 [24] |
| Oral squamous cell carcinoma | Upregulated | Oncogenic | not investigated | Apoptosis, migration, invasion | miR-185-5p/Rab14 [25] |
| Tongue squamous cell carcinoma | Upregulated | Oncogenic | Clinical stage, node metastasis, survival status, cisplastin sensitivity | Proliferation, migration, invasion, cisplatin resistance | miR-211-5p/Ezrin/Fak/Src [26]; miR-124-3p/TRIM14 [7] |
| Maxillary sinus squamous cell carcinoma | Upregulated | Oncogenic | not investigated | Viability, migration, invasion, | miR-204/EphA7 [27] |
| Acute myeloid leukemia | Upregulated | Oncogenic | not investigated | Proliferation, apoptosis, PMA-induced differentiation | miR-326/c-Myc [28] |
| Osteosarcoma | Upregulated | Oncogenic | not investigated | Proliferation, apoptosis, migration, invasion, EMT, aerobic gylcosis, fluorouracil resistance, | β-catenin [29]; KCNQ1/DNMT1 [30]; miR-4458/CCND2 [31]; miR-34c-5p/ALDOA [1]; miR-192-5p/LARP1 [2] |
| Chordoma | Upregulated | Oncogenic | not investigated | Multidrug resistance | miR-27b-3p/ATF2 [4] |
| Breast cancer | Upregulated | Oncogenic | Tumor size, tumor count, tumor stage | Proliferation, cell cycle, apoptosis, migration | miR-145/CCNE2 [5]; miR-107 [6] |
| Cholangiocarcinoma | Upregulated | Oncogenic | Tumor site, differentiation grade, tumor stage, TMN stage, lymph node metastasis, postoperative recurrence | Proliferation, apoptosis, invasion, EMT | miR-140-5p/SOX4 [8] |
| Hepatocellular carcinoma | Upregulated | Oncogenic | not investigated | Proliferation, viability, survival, apoptosis, migration, invasion, metastasis, oxaliplatin and sorafenib resistance | miR-7-5p/ABCC1 [9]; miR-149/S1PR1/PI3K/AKT [10]; miR-506/PD-L1 [11]; miR-148a-3p/IGF1R [12] |
| Bladder cancer | Upregulated | Oncogenic | Poor prognosis | Proliferation, apoptosis, migration, invasion | miR-145-5p/PCBP2 [32] |
| Lung cancer | Upregulated | Oncogenic | Tumor size, TNM stage, disease stage, lymph node metastasis, histological differentiation, smoking history, OS | Proliferation, cell cycle, autophagy, apoptosis, migration, invasion, aerobic glycolysis, multidrug resistance, irradiation resistance | miR-204-5p/ATG3 [33]; MDR1 [34]; miR-27b-3p/HSP90AA1 [35]; miR-129-5p/JAG1 [36]; miR-372-3p/ATG5/ATG12 [37]; JAK2/STAT3 [38]; TGF-β1 [39] |
| Lung cancer | Upregulated | Suppressor | Clinical stage, tumor size, lymph node metastasis | Proliferation | not investigated [40] |
| Melanoma | Upregulated | Oncogenic | Poor prognosis | Proliferation, metastasis | miR-153/MET [41] |
| Prostate cancer | Upregulated | Oncogenic | not investigated | Proliferation, apoptosis, migration, invasion, metastasis | miR-15a/PD-L1 [42]; miR-211-5p/CHI3L1 [43] |
| Gastric cancer | Upregulated/Downregulated | Oncogenic/Suppressor | TNM stage, local invasion, lymph node metastasis, distant metastasis, histological grade | Proliferation, viability, survival, apoptosis, migration, invasion | miR-9/LMX1A [44]; miR-145-5p/ARF6 [45]; miR-43198/DRAM2 [46] |
Key †: TNM (tumor lymph node metastasis), overall survival (OS), disease-free survival (DFS), epithelial–mesenchymal transition (EMT).
3. Regulatory Mechanisms of KCNQ1OT1
3.1. Impact of KCNQ1OT1 on microRNA Regulation
LncRNAs acting as ceRNAs with miRNAs are the most frequently identified biological function of lncRNAs and has been associated with various cancers, such as lung [33][47], prostate [48], ovarian [20][49], colorectal [50], and glioblastoma multiforme [51]. By sponging miR-217, KCNQ1OT1 upregulated zinc finger E-box binding homeobox 1 (ZEB1) and regulated CRC cell proliferation, migration, and EMT formation [13] (Table 1). Similarly, the miR-329-3p/CTNND1 (Catenin delta-1) axis was demonstrated to interact with KCNQ1OT1 to modulate SW480 and LS1034 CRC cancer cell proliferation, migration, invasion, and apoptosis [15]. In a xenograft mouse model, knockdown of KCNQ1OT1 inhibited CRC cell growth and decreased tumor volume, while overexpression of KCNQ1OT1 induced protective autophagy and chemoresistance to oxaliplatin (OXA) by sponging miR-34a and upregulating autophagy-related 4B (Atg4B) [14]. Sun et al., showed that KCNQ1OT1 acted as a sponge of miR-204 in the progression of MSSCC and explored the role of ceRNA regulation of the KCNQ1OT1/miR-204/EphA7 axis [27]. Cell-based studies indicate that KCNQ1OT1 knockdown inhibited cell proliferation and promoted apoptosis and cell differentiation in HL-60 and U937 AML cells by acting as a ceRNA for miR-326 and targeting c-Myc (Myc proto-oncogene, basic helix-loop-helix (bHLH) transcription factor) [28]. Loss of KCNQ1OT1 inhibited BRCA cell proliferation and migration in BT-549 and HCC1599 cells and reduced tumor growth in vivo by sponging miR-107 [6] (Table 1). Additional experiments are needed to investigate whether cyclin-dependent kinase 8 (CDK8) is involved in the epigenetic regulation of the KCNQ1OT1–hsa-miR-107 axis, as CDK8 has been previously shown by Li et al. to regulate miR-107 in BRCA [52].
KCNQ1OT1 serves as ceRNA to regulate multidrug resistance via regulating miR-27b-3p/activating transcription factor 2 (ATF2) in human chordoma bone tumor cells [4]. Zhu et al. hypothesized that KCNQ1OT1 is correlated with poor prognosis in patients with soft tissue sarcoma (STS), competitively binds with miR-29c-3p, regulating JARID2, CDK6, DNMT3A, and TET [24] (Table 1). Thus, this intricate ceRNA network may serve as a therapeutic target for treating the STS sub-cluster of patients with a poor prognosis. KCNQ1OT1 upregulation induced cell proliferation and migration and inhibited apoptosis in HOS and U2OS OS cells through competitive binding of miR-4458 and upregulating cyclin D2 (CCND2) [31]. KCNQ1OT1 also promoted U2OS and 143B OS cell proliferation by enhancing aerobic glycolysis through competitive binding to miR-34c-5p and stimulating aldolase A (ALDOA) expression in vitro and in vivo [1] (Table 1).
KCNQ1OT1 acts as a competing endogenous RNA (ceRNA) for miR-145-5p, resulting in increased expression of poly(rC)-binding protein 2 (PCBP2). PCBP2 is a target of miR-145-5p, and its overexpression results in the progression of BC by modulating cell proliferation, migration and invasion, and cell apoptosis [32] (Table 1). Therefore, KCNQ1OT1 expression may identify the subset of BC patients with a more aggressive phenotype. Wang et al., have shown that KCNQ1OT1 also sponged miR-129-5p and regulated jagged canonical Notch ligand 1 (JAG1) expression that induces proliferation, migration, and invasion of A549 and H460 NSCLC cells [36] (Table 1). KCNQ1OT1 was upregulated in irradiation-resistant LAD cells and is associated with the low response to anticancer treatment and poor prognosis of LAD patients [37]. Knockdown in stereotactic body radiation therapy-resistant cells significantly enhanced radiosensitivity both in vitro and in vivo by sponging miR-372-3p and regulating autophagy-related targets (ATG5 and ATG12), thereby inhibiting autophagy (Table 1). KCNQ1OT1 is aberrantly upregulated in melanoma and retinoblastoma (RB) patient tissues compared with adjacent normal tissues [41][53][54]. Overexpression of KCNQ1OT1 contributed to the proliferation, migration, and invasion of melanoma and RB cells by sponging miR-153, and increasing MET proto-oncogene receptor tyrosine kinase (MET) and hypoxia-inducible factor-1α (HIF-1α) expression, respectively [41][53] (Table 1). KCNQ1OT1 also acts as a ceRNA for miR-124 to promote RB cell progression by regulating the transcription factor, specificity protein 1 (SP1) expression, and the silent information regulator 1 (SIRT1)/c-Jun N-terminal kinase (JNK) signaling pathway [54].
KCNQ1OT1 was highly expressed in CRC, and its knockdown suppressed cell proliferation, migration, and invasion by interacting with miR145-5p/zinc finger protein 146 (ZNF146) [16]. In addition, Wang et al. found that KCNQ1OT1 promoted GC progression by sponging miR-4319 to upregulate the expression of DNA-damage-regulated autophagy modulator 2 (DRAM2) [46]. Moreover, KCNQ1OT1 is discussed as a ceRNA for miR-148a-3p and a positive regulator for IGF1R in HCC [12].
In a recent study by Chen et al., KCNQ1OT1, PD-L1, and CD8 levels were significantly increased in prostate cancer tissues compared with adjacent non-tumor tissues [42]. KCNQ1OT1 was shown to regulate PD-L1 expression by sponging miR-15a in PCa, resulting in the inhibition of cytotoxicity of CD8+ T cells and promotion of tumor evasion (Table 1). Furthermore, knockdown of KCNQ1OT1 significantly decreased PD-L1 expression, inhibited the viability, migration, invasion, and EMT, promoted apoptosis of PCa cells, and enhanced the function of CD8+ T cells. Zhang et al., further demonstrated that knockdown of KCNQ1OT1 could reduce sorafenib resistance and PD-L1-mediated immune escape, regulate cytokine secretion and CD8+ T-cell apoptosis, and suppress migration and invasion in sorafenib-resistant HCC cells by sponging miR-506 [11]. These findings indicate that KCNQ1OT1 plays a significant oncogenic role in PCa and HCC tumorigenesis and may become a promising therapy that targets tumor evasion and drug resistance and inhibits the malignant growth of cells.
KCNQ1OT1 expression was positively associated with Chitinase 3 Like 1 (CHI3L1) expression and significantly promotes prostate cancer (PCa) cell proliferation, invasion, and metastasis [43]. Hao et al. found that overexpression of KCNQ1OT1 competes with miR-211-5p expression, which functions as a ceRNA to promote CHI3L1 expression and PCa progression [43] (Table 1). These results suggest that KCNQ1OT1 may be a prognostic marker for poor outcomes in PCa. In contrast, Li et al., reported that overexpression of KCNQ1OT1 promoted apoptosis in neuroblastoma cells by sponging miR296-5p and upregulating BCL2 Associated X (Bax), a key regulator of cell death [23], suggesting a cancer cell type-dependent role of KCNQ1OT1.
3.2. Impact of KCNQ1OT1 on Cell Signaling Pathways
An increasing number of studies have demonstrated that lncRNAs modulate oncogenic signaling [55][56], highlighting their utility as diagnostic markers and therapeutic targets [25]. Several studies have revealed that KCNQ1OT1 is overexpressed in SCLC patients and is associated with a poor prognosis. Downregulation of KCNQ1OT1 inhibits SCLC cell proliferation, migration, and invasion, induces apoptosis, and suppresses tumor growth and chemoresistance via TGF-β-mediated EMT signaling [49] and the Janus kinase (JAK)/signal transducer and activator of transcription 3 (STAT3) signaling pathway [48]. Duan et al. reported that KCNQ1OT1 promoted the malignancy SW620 and RKO CRC cells by upregulation of the PI3K/AKT signaling pathway [34]. Moreover, KCNQ1OT1 promoted OS cell proliferation, migration, invasion, and EMT in primary osteosarcoma cells by activating β-catenin [57].
This entry is adapted from the peer-reviewed paper 10.3390/biom11111602
References
- Shen, Y.; Xu, J.; Pan, X.; Zhang, Y.; Weng, Y.; Zhou, D.; He, S. LncRNA KCNQ1OT1 sponges miR-34c-5p to promote osteosarcoma growth via ALDOA enhanced aerobic glycolysis. Cell Death Dis. 2020, 11, 278.
- Zhang, Y.; Cai, W.; Zou, Y.; Zhang, H. Knockdown of KCNQ1OT1 Inhibits Proliferation, Invasion, and Drug Resistance by Regulating miR-129-5p-Mediated LARP1 in Osteosarcoma. BioMed Res. Int. 2020, 2020, 7698767.
- Xian, D.; Zhao, Y. LncRNA KCNQ1OT1 enhanced the methotrexate resistance of colorectal cancer cells by regulating miR-760/PPP1R1B via the cAMP signalling pathway. J. Cell Mol. Med. 2019, 23, 3808–3823.
- Li, L.; Lv, G.; Wang, B.; Ma, H. Long Non-Coding RNA KCNQ1OT1 Promotes Multidrug Resistance in Chordoma by Functioning as a Molecular Sponge of miR-27b-3p and Subsequently Increasing ATF2 Expression. Cancer Manag. Res. 2020, 12, 7847–7853.
- Feng, W.; Wang, C.; Liang, C.; Yang, H.; Chen, D.; Yu, X.; Zhao, W.; Geng, D.; Li, S.; Chen, Z.; et al. The Dysregulated Expression of KCNQ1OT1 and Its Interaction with Downstream Factors miR-145/CCNE2 in Breast Cancer Cells. Cell. Physiol. Biochem. 2018, 49, 432–446.
- Wu, Y.; Bi, Q.J.; Han, R.; Zhang, Y. Long noncoding RNA KCNQ1OT1 is correlated with human breast cancer cell development through inverse regulation of hsa-miR-107. Biochem. Cell Biol. 2020, 98, 338–344.
- Qiao, C.Y.; Qiao, T.Y.; Jin, H.; Liu, L.L.; Zheng, M.D.; Wang, Z.L. LncRNA KCNQ1OT1 contributes to the cisplatin resistance of tongue cancer through the KCNQ1OT1/miR-124-3p/TRIM14 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 200–212.
- Sun, H.; Li, Y.; Kong, H.; Dai, S.; Qian, H. Dysregulation of KCNQ1OT1 promotes cholangiocarcinoma progression via miR-140-5p/SOX4 axis. Arch. Biochem. Biophys. 2018, 658, 7–15.
- Hu, H.; Yang, L.; Li, L.; Zeng, C. Long non-coding RNA KCNQ1OT1 modulates oxaliplatin resistance in hepatocellular carcinoma through miR-7-5p/ ABCC1 axis. Biochem. Biophys. Res. Commun. 2018, 503, 2400–2406.
- Cheng, J.L.; Li, D.J.; Lv, M.Y.; Pei, Y.J.; Zhang, X.J.; Li, L.; Liu, X.Y.; Fan, A.H. LncRNA KCNQ1OT1 regulates the invasion and migration of hepatocellular carcinoma by acting on S1PR1 through miR-149. Cancer Gene Ther. 2020.
- Zhang, J.; Zhao, X.; Ma, X.; Yuan, Z.; Hu, M. KCNQ1OT1 contributes to sorafenib resistance and programmed death-ligand-1-mediated immune escape via sponging miR-506 in hepatocellular carcinoma cells. Int. J. Mol. Med. 2020, 46, 1794–1804.
- Xu, G.; Zhu, Y.; Liu, H.; Liu, Y.; Zhang, X. Long Non-Coding RNA KCNQ1OT1 Promotes Progression of Hepatocellular Carcinoma by miR-148a-3p/IGF1R Axis. Technol. Cancer Res. Treat. 2020, 19, 1533033820980117.
- Bian, Y.; Gao, G.; Zhang, Q.; Qian, H.; Yu, L.; Yao, N.; Qian, J.; Liu, B.; Qian, X. KCNQ1OT1/miR-217/ZEB1 feedback loop facilitates cell migration and epithelial-mesenchymal transition in colorectal cancer. Cancer Biol. Ther. 2019, 20, 886–896.
- Li, Y.; Li, C.; Li, D.; Yang, L.; Jin, J.; Zhang, B. lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR-34a/ATG4B pathway. Onco Targets Ther. 2019, 12, 2649–2660.
- Liu, X.; Zhang, Y.; Wang, Y.; Bian, C.; Wang, F. Long non-coding RNA KCNQ1OT1 up-regulates CTNND1 by sponging miR-329-3p to induce the proliferation, migration, invasion, and inhibit apoptosis of colorectal cancer cells. Cancer Cell Int 2020, 20, 340.
- Zhu, S.; Chen, C.-Y.; Hao, Y. LncRNA KCNQ1OT1 acts as miR-216b-5p sponge to promote colorectal cancer progression via up-regulating ZNF146. J. Mol. Histol. 2021, 52, 479–490.
- Chen, C.; Wei, M.; Wang, C.; Sun, D.; Liu, P.; Zhong, X.; Yu, W. Long noncoding RNA KCNQ1OT1 promotes colorectal carcinogenesis by enhancing aerobic glycolysis via hexokinase-2. Aging (Albany NY) 2020, 12, 11685–11697.
- Duan, Q.; Cai, L.; Zheng, K.; Cui, C.; Huang, R.; Zheng, Z.; Xie, L.; Wu, C.; Yu, X.; Yu, J. lncRNA KCNQ1OT1 knockdown inhibits colorectal cancer cell proliferation, migration and invasiveness via the PI3K/AKT pathway. Oncol. Lett. 2020, 20, 601–610.
- Lin, Z.-B.; Long, P.; Zhao, Z.; Zhang, Y.-R.; Chu, X.-D.; Zhao, X.-X.; Ding, H.; Huan, S.-W.; Pan, Y.-L.; Pan, J.-H. Long Noncoding RNA KCNQ1OT1 is a Prognostic Biomarker and mediates CD8(+) T cell exhaustion by regulating CD155 Expression in Colorectal Cancer. Int. J. Biol. Sci. 2021, 17, 1757–1768.
- Liu, H.; Chen, R.; Kang, F.; Lai, H.; Wang, Y. KCNQ1OT1 promotes ovarian cancer progression via modulating MIR-142-5p/CAPN10 axis. Mol. Genet. Genom. Med. 2020, 8, e1077.
- Lei, H.W.; Gao, Y.; Shi, J.B.; Teng, Y.; Song, C.H.; Zou, L.J.; Ye, F.X.; Zhang, H.C. KCNQ1 opposite strand/antisense transcript 1 promotes aggressive biological behaviors of cervical cancer cells via regulating microRNA-491-5p and pyruvate kinase M1/2. J. Biol. Regul. Homeost. Agents 2020, 34.
- Gong, W.; Zheng, J.; Liu, X.; Liu, Y.; Guo, J.; Gao, Y.; Tao, W.; Chen, J.; Li, Z.; Ma, J.; et al. Knockdown of Long Non-Coding RNA KCNQ1OT1 Restrained Glioma Cells’ Malignancy by Activating miR-370/CCNE2 Axis. Front. Cell Neurosci. 2017, 11, 84.
- Li, M.M.; Liu, X.H.; Zhao, Y.C.; Ma, X.Y.; Zhou, Y.C.; Zhao, Y.X.; Liu, X.Y. Long noncoding RNA KCNQ1OT1 promotes apoptosis in neuroblastoma cells by regulating miR-296-5p/Bax axis. FEBS J. 2020, 287, 561–577.
- Zhu, Z.; Jin, Z.; Zhang, H.; Zhang, M.; Sun, D. Integrative Clustering Reveals a Novel Subtype of Soft Tissue Sarcoma With Poor Prognosis. Front. Genet. 2020, 11, 69.
- Bao, Q.; Liao, X.; Li, R.; Ding, N. KCNQ1OT1 promotes migration and inhibits apoptosis by modulating miR-185-5p/Rab14 axis in oral squamous cell carcinoma. Dev. Growth Differ. 2019, 61, 466–474.
- Zhang, S.; Ma, H.; Zhang, D.; Xie, S.; Wang, W.; Li, Q.; Lin, Z.; Wang, Y. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018, 9, 742.
- Sun, Y.; Xu, C.; Wu, Q.; Zhang, L.; Wang, P. Long noncoding RNA KCNQ1OT1 promotes proliferation, migration, and invasion in maxillary sinus squamous cell carcinoma by regulating miR-204/EphA7 axis. J. Cell Biochem. 2020, 121, 2962–2969.
- Cheng, P.; Lu, P.; Guan, J.; Zhou, Y.; Zou, L.; Yi, X.; Cheng, H. LncRNA KCNQ1OT1 controls cell proliferation, differentiation and apoptosis by sponging miR-326 to regulate c-Myc expression in acute myeloid leukemia. Neoplasma 2020, 67, 238–248.
- Zhang, C.; Du, S.; Cao, L. Long non-coding RNA KCNQ1OT1 promotes osteosarcoma progression by increasing β-catenin activity. RSC Adv. 2018, 8, 37581–37589.
- Qi, X.; Yu, X.-J.; Wang, X.-M.; Song, T.-N.; Zhang, J.; Guo, X.-Z.; Li, G.-J.; Shao, M. Knockdown of KCNQ1OT1 Suppresses Cell Invasion and Sensitizes Osteosarcoma Cells to CDDP by Upregulating DNMT1-Mediated Kcnq1 Expression. Mol. Ther.–Nucleic Acids 2019, 17, 804–818.
- Wang, M.; Wang, Z.; Zhu, X.; Guan, S.; Liu, Z. LncRNA KCNQ1OT1 acting as a ceRNA for miR-4458 enhances osteosarcoma progression by regulating CCND2 expression. In Vitro Cell Dev. Biol. Anim. 2019, 55, 694–702.
- Wang, J.; Zhang, H.; Situ, J.; Li, M.; Sun, H. KCNQ1OT1 aggravates cell proliferation and migration in bladder cancer through modulating miR-145-5p/PCBP2 axis. Cancer Cell Int. 2019, 19, 325.
- Kang, Y.; Jia, Y.; Wang, Q.; Zhao, Q.; Song, M.; Ni, R.; Wang, J. Long Noncoding RNA KCNQ1OT1 Promotes the Progression of Non-Small Cell Lung Cancer via Regulating miR-204-5p/ATG3 Axis. Onco Targets Ther. 2019, 12, 10787–10797.
- Ren, K.; Xu, R.; Huang, J.; Zhao, J.; Shi, W. Knockdown of long non-coding RNA KCNQ1OT1 depressed chemoresistance to paclitaxel in lung adenocarcinoma. Cancer Chemother. Pharmacol. 2017, 80, 243–250.
- Dong, Z.; Yang, P.; Qiu, X.; Liang, S.; Guan, B.; Yang, H.; Li, F.; Sun, L.; Liu, H.; Zou, G.; et al. KCNQ1OT1 facilitates progression of non-small-cell lung carcinoma via modulating miRNA-27b-3p/HSP90AA1 axis. J. Cell Physiol. 2019, 234, 11304–11314.
- Wang, Y.; Zhang, L.; Yang, J.; Sun, R. LncRNA KCNQ1OT1 promotes cell proliferation, migration and invasion via regulating miR-129-5p/JAG1 axis in non-small cell lung cancer. Cancer Cell Int. 2020, 20, 144.
- He, H.; Song, X.; Yang, Z.; Mao, Y.; Zhang, K.; Wang, Y.; Su, B.; Li, Q.; Chen, H.; Li, Y. Upregulation of KCNQ1OT1 promotes resistance to stereotactic body radiotherapy in lung adenocarcinoma by inducing ATG5/ATG12-mediated autophagy via miR-372-3p. Cell Death Dis. 2020, 11, 883.
- Zhu, Y.; Shen, Y.; Chen, R.; Li, H.; Wu, Y.; Zhang, F.; Huang, W.; Guo, L.; Chen, Q.; Liu, H. KCNQ1OT1 lncRNA affects the proliferation, apoptosis, and chemoresistance of small cell lung cancer cells via the JAK2/STAT3 axis. Ann. Transl. Med. 2021, 9, 891.
- Li, D.; Tong, Q.; Lian, Y.; Chen, Z.; Zhu, Y.; Huang, W.; Wen, Y.; Wang, Q.; Liang, S.; Li, M.; et al. Inhibition of lncRNA KCNQ1OT1 Improves Apoptosis and Chemotherapy Drug Response in Small Cell Lung Cancer by TGF-β1 Mediated EMT. J. Korean Cancer Assoc. 2021, 53, 1042–1056.
- Sun, X.; Xin, Y.; Wang, M.; Li, S.; Miao, S.; Xuan, Y.; Wang, Y.; Lu, T.; Liu, J.; Jiao, W. Overexpression of long non-coding RNA KCNQ1OT1 is related to good prognosis via inhibiting cell proliferation in non-small cell lung cancer. Thorac. Cancer 2018, 9, 523–531.
- Guo, B.; Zhang, Q.; Wang, H.; Chang, P.; Tao, K. KCNQ1OT1 promotes melanoma growth and metastasis. Aging (Albany NY) 2018, 10, 632–644.
- Chen, Q.-H.; Li, B.; Liu, D.-G.; Zhang, B.; Yang, X.; Tu, Y.-L. LncRNA KCNQ1OT1 sponges miR-15a to promote immune evasion and malignant progression of prostate cancer via up-regulating PD-L1. Cancer Cell Int. 2020, 20, 394.
- Hao, H.; Chen, H.; Xie, L.; Liu, H.; Wang, D. LncRNA KCNQ1OT1 Promotes Proliferation, Invasion and Metastasis of Prostate Cancer by Regulating miR-211-5p/CHI3L1 Pathway. Onco Targets Ther. 2021, 14, 1659–1671.
- Feng, L.; Li, H.; Li, F.; Bei, S.; Zhang, X. LncRNA KCNQ1OT1 regulates microRNA-9-LMX1A expression and inhibits gastric cancer cell progression. Aging (Albany NY) 2020, 12, 707–717.
- Zhong, X.; Wen, X.; Chen, L.; Gu, N.; Yu, X.; Sui, K. Long non-coding RNA KCNQ1OT1 promotes the progression of gastric cancer via the miR-145-5p/ARF6 axis. J. Gene Med. 2021, 23, e3330.
- Wang, J.; Wu, F.; Li, Y.; Pang, L.; Wang, X.; Kong, G.; Zhang, T.; Yu, D. KCNQ1OT1 accelerates gastric cancer progression via miR-4319/DRAM2 axis. Int. J. Immunopathol. Pharmacol. 2020, 34, 2058738420954598.
- Ginn, L.; Shi, L.; Montagna, M.; Garofalo, M. LncRNAs in Non-Small-Cell Lung Cancer. Noncoding RNA 2020, 6, 25.
- Xu, Y.-H.; Deng, J.-L.; Wang, G.; Zhu, Y.-S. Long non-coding RNAs in prostate cancer: Functional roles and clinical implications. Cancer Lett. 2019, 464, 37–55.
- Zhou, M.; Wang, X.; Shi, H.; Cheng, L.; Wang, Z.; Zhao, H.; Yang, L.; Sun, J. Characterization of long non-coding RNA-associated ceRNA network to reveal potential prognostic lncRNA biomarkers in human ovarian cancer. Oncotarget 2016, 7, 12598–12611.
- Wang, L.; Cho, K.B.; Li, Y.; Tao, G.; Xie, Z.; Guo, B. Long Noncoding RNA (lncRNA)-Mediated Competing Endogenous RNA Networks Provide Novel Potential Biomarkers and Therapeutic Targets for Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 5758.
- Zhang, K.; Li, Q.; Kang, X.; Wang, Y.; Wang, S. Identification and functional characterization of lncRNAs acting as ceRNA involved in the malignant progression of glioblastoma multiforme. Oncol Rep. 2016, 36, 2911–2925.
- Li, X.-Y.; Luo, Q.-F.; Wei, C.-K.; Li, D.-F.; Li, J.; Fang, L. MiRNA-107 inhibits proliferation and migration by targeting CDK8 in breast cancer. Int. J. Clin. Exp. Med. 2014, 7, 32–40.
- Wang, Y.; Wang, J.; Hao, H.; Luo, X. lncRNA KCNQ1OT1 promotes the proliferation, migration and invasion of retinoblastoma cells by upregulating HIF-1α via sponging miR-153-3p. J. Investig Med. 2020, 68, 1349–1356.
- Zhang, H.; Yang, X.; Xu, Y.; Li, H. KCNQ1OT1 regulates the retinoblastoma cell proliferation, migration and SIRT1/JNK signaling pathway by targeting miR-124/SP1 axis. Biosci. Rep. 2021, 41, BSR20201626.
- Fu, P.-F.; Zheng, X.; Fan, X.; Lin, A.-F. Role of cytoplasmic lncRNAs in regulating cancer signaling pathways. J. Zhejiang Univ. Sci. B 2019, 20, 1–8.
- Peng, W.X.; Koirala, P.; Mo, Y.Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667.
- Jia, Z.W.; Li, Y.; Cui, G.R.; Zhao, H.B.; Li, P.Y.; Luo, J.M. Expression and Clinical Significance of LncRNA KCNQ1OT1 in Patients with Acute Myeloid Leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2018, 26, 653–657.
This entry is offline, you can click here to edit this entry!
