Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biochemistry & Molecular Biology
Persulfidation is a post-translational modification (PTM) of proteins that affects the thiol group (-SH) of some cysteine residues (Cys) which could modify either positively or negatively the function of the target protein.
- Hydrogen Sulfide
- Protein Persulfidation
1. Introduction
Hydrogen sulfide (H2S) is currently recognized as a new signaling molecule, with cytoprotective properties similar to those of gasotransmitters such as nitric oxide (NO) and carbon monoxide (CO).
2. Protein Persulfidation and Other Cysteine Modifications
Over the last ten years, the development of new mass spectrometry (MS)-based high-throughput proteomic techniques has led to an increase in the identification of proteins susceptible to persulfidation. In spinach (Spinacia oleracea) plants exposed to 100 µM sodium hydrosulfide (NaHS), an H2S donor, proteomic analysis of leaves has shown how H2S affects protein expression. Thus, of the 1000 proteins identified, 92 were differentially expressed (an increase of over 2-fold), 65 were up-regulated and 27 down-regulated due to the treatment [1]. On the other hand, using the biotin switch method, suitably adapted in order to identify persulfidated proteins, combined with liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, a total of 106 putative proteins targeted for persulfidation were identified in Arabidopsis thaliana leaves [2]. Subsequently, using an improved tag-switch method, these authors expanded the number of persulfidated proteins to 2015 [3]. More recently, the number of persulfidated proteins in arabidopsis roots has reached 5214 through the use of extremely high-resolution mass spectrometer [4]. These plant proteomic studies that identified potential protein targets of persulfidation were used as the starting point for complementary analyses to specifically identify the residue(s) persulfidated and to determine the effect of this oxiPTM on the function of the proteins targeted [5]. Table 1 [6][7][8][9][10][11][12][13][14][15][16][17][18] shows an updated list of the plant proteins undergoing persulfidation, and the effect on protein function. These persulfidated proteins are involved in a wide range of cellular processes, including photosynthesis, the sulfur metabolism, reactive oxygen/nitrogen species (ROS/RNS) activity, ethylene biosynthesis, organelle movements, autophagy and ABA signaling, thus confirming the important role of H2S in plant cell physiology.
Table 1. Identified plant protein targets of persulfidation whose function is positive or negatively affected by H2S.
| Enzyme/Protein | Function | Effect | Ref. |
|---|---|---|---|
| RuBISCO | Photosynthesis | Activity up-regulated | [6] |
| O-acetylserine(thiol)lyase (OAS-TL) | Sulfur metabolism | Activity up-regulated | [6] |
| L-cysteine desulphydrase (LCD) | Sulfur metabolism | Activity up-regulated | [7] |
| Ascorbate peroxidase (APX) | Antioxidant | Activity up-regulated | [19][2] |
| Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) | Production of energy in the glycolysis | Activity up-regulated | [2] |
| Glutamine synthetase (GS) | Metabolism of nitrogen | Activity down-regulated | [2] |
| Actin | Organelle movement, cell division and expansion | Inhibits actin polymerization | [8] |
| 1-aminocyclopropane-1-carboxylic acid oxidase (ACO) | Ethylene biosynthesis | Activity down-regulated | [9] |
| NADP-isocitrate dehydrogenase (NADP-ICDH) | Provides NADPH as a reducing agent | Activity down-regulated | [10] |
| NADP-malic enzyme (NADP-ME) | Provides NADPH as a reducing agent | Activity down-regulated | [11] |
| Catalase | Antioxidant | Activity down-regulated | [20][19] |
| SNF1-RELATED PROTEIN KINASE2.6 (SnRK2.6) | Promotes ABA signaling | Promotes stomatal closureActivity up-regulated | [12][13] |
| Respiratory burst oxidase homolog protein D (RBOHD) | Generation of superoxide radical | Activity up-regulated | [7] |
| Cysteine protease ATG4 | Autophagy | Inhibits autophagy | [14] |
| ATG18 | Autophagy | Negative regulation | [15] |
| Peroxidase | ROS metabolism | Activity up-regulated | [19] |
| Flowering Locus C protein (FLC1 and 3) | Flowering regulatory pathway | Reduces binding abilities of FLCs | [16] |
| Mitogen-activated protein kinase (MPK) 4 | Allivates cold stress | Activity up-regulated | [17] |
| Abscisic acid insensitive 4 (ABI4) | Regulates ABA signaling | Enhanced the transactivation activity of ABI4 towards MAPKKK18 | [18] |
It is worth noting that, following NaHS treatment, the H2S-generating enzyme, L-cysteine desulfhydrase1 (DES1), is persulfidated in Cys44 and Cys205 and subsequently upregulated [7]. Likewise, respiratory burst oxidase homolog protein D (RBOHD), which is involved in stomatal movements, is also persulfidated in Cys825 and Cys890 and its activity is upregulated [7].
In Arabidopsis thaliana, 3-mercaptopyruvate sulfurtransferase (MST), specifically the sulfurtransferase 1 and sulfurtransferase 2 (STR1 and STR2), has recently been shown to mediate either protein persulfidation or H2S formation, as well as low molecular weight persulfide formation [21]. S-desulfurization, a new GSH-mediated process, as opposed to persulfidation, involving the release of H2S from persulfidated proteins, has also been reported. S-desulfurization mitigates the inhibition by persulfidation of various enzymes including alliinase, D-lactate dehydrogenase (D-LDH), alcohol dehydrogenase (ADH) and glucose-6-phosphate dehydrogenase (G6PDH) [22]. These findings provide further confirmation of the important role played by persulfidation in the regulation of its target proteins.
In addition to persulfidation, other competing PTMs can affect thiol groups. Figure 1 shows a summary of the principal oxiPTMs in which protein thiol groups are involved. S-glutathionylation is mediated by the addition of the tripeptide glutathione (GSH) γ-L-glutamyl-L-cysteinylglycine [23], an antioxidant molecule which is one of the most abundant low molecular mass thiols in plant cells [24]. GSH also interacts with NO to form S-nitrosoglutathtione (GSNO) which is regarded as a NO reservoir [25]. S-sulfenylation, which results from the oxidation of the thiol group to Cys sulfenic acid, is mediated by H2O2 [26][27]. S-acylation is a reversible oxiPTM involving the addition of a fatty acid, such as palmitate or stearate, to specific cysteines through thioester bonds [28][29]. S-cyanylation is a new PTM mediated by cyanide (CN-) [30] which opens up a whole new range of functions that need to be explored. Recently, an in silico platform named pCysMod was set up to predict multiple Cys PTMs [31], such as S-nitrosation, S-glutathionylation, S-cyanylation, S-sulfenylation and S-acylation, due to interactions with NO, GSH, cyanide, hydrogen peroxide (H2O2) and fatty acids, respectively. The integrative database iCysMod (http://icysmod.omicsbio.info/#; accessed on 26 October 2021) for protein cysteine modifications (PCMs) in 48 eukaryotes including Arabidopsis thaliana has also been established [32]. This database is a useful tool for investigating the mechanisms underlying the regulatory processes involving these oxiPTMs.
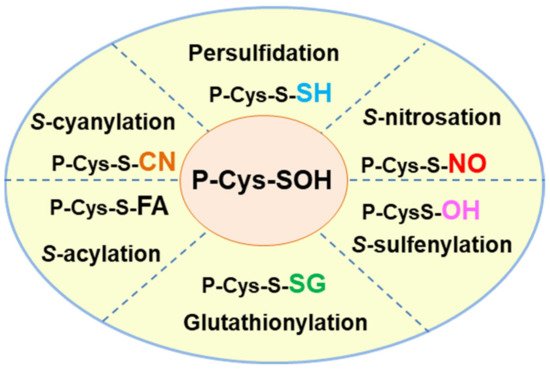
Figure 1. Hallmark of the different oxidative posttranslational modifications (oxiPTMs) of cysteine-containing proteins including persulfidation, S-nitrosation, S-sulfenylation, glutathionylation, S-acylation and S-cyanylation which are mediated by hydrogen sulfide (H2S), nitric oxide (NO), hydrogen peroxide (H2O2), glutathione (GSH), fatty acid (FA) and cyanide (CN-), respectively. The thiol group (-SH) of Cys residues works as a redox switch and each of the PTMs can modify the function of the target protein either positively or negatively.
In higher plants, ascorbate peroxidase (APX) constitutes a good example of a protein whose Cys residues are affected by oxiPTMs. This key antioxidant enzyme, which is present in virtually all subcellular compartments, is a component in the ascorbate-glutathione cycle [33][34][35][36][37]. APX catalyzes the decomposition of H2O2 associated with the oxidation of ascorbate to dehydroascorbate. Various studies have shown that, in a diverse range of plant species, APX is targeted by oxiPTMs including glutathionylation [36], S-nitrosation [38][39] (Begara-Morales et al. 2014; Hu et al. 2015), S-persulfidation [2], S-sulfenylation [26] and S-cyanylation [30] (García et al., 2019). While APX activity is prone to sulfenylation involving inactivation by critical cysteine oxidation, the other PTMs appear to act protectively. Catalase (CAT), the principal H2O2-catalyzing enzyme in eukaryotes, is also susceptible to post-translational modification by oxidation, nitration, S-nitrosylation and persulfidation [20][40]. CAT, which, with its high Km reaction rate, dismutates hydrogen peroxide, as well as APX, which fine-tunes CAT’s enzymatic role, acts synergistically to control H2O2 levels. This diversity of regulatory events points to how all these oxyPTMs modulate H2O2 content in different subcellular compartments, particularly in peroxisomes, where CAT and APX are present. There are other examples such as some NADPH generating enzymes, i.e., NADP-glyceraldehyde-3-phosphate dehydrogenase and NADP-isocitrate dehydrogenase, which can undergo some of these oxiPTMs (persulfidation and S-nitrosation) affecting its capacity to generate NADPH and, consequently, modulate the redox state of the cell [41].
This entry is adapted from the peer-reviewed paper 10.3390/antiox10111686
References
- Chen, J.; Liu, T.W.; Hu, W.J.; Simon, M.; Wang, W.H.; Chen, J.; Liu, X.; Zheng, H.L. Comparative proteomic analysis of differentially expressed proteins induced by hydrogen sulfide in Spinacia oleracea leaves. PLoS ONE 2014, 9, e105400.
- Aroca, Á.; Serna, A.; Gotor, C.; Romero, L.C. S-sulfhydration: A cysteine posttranslational modification in plant systems. Plant Physiol. 2015, 168, 334–342.
- Aroca, A.; Benito, J.M.; Gotor, C.; Romero, L.C. Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J. Exp. Bot. 2017, 68, 4915–4927.
- Jurado-Flores, A.; Romero, L.C.; Gotor, C. Label-Free quantitative proteomic analysis of nitrogen starvation in arabidopsis root reveals new aspects of H2S signaling by protein persulfidation. Antioxidants 2021, 10, 508.
- Zhang, J.; Zhou, M.; Zhou, H.; Zhao, D.; Gotor, C.; Romero, L.C.; Shen, J.; Ge, Z.; Zhang, Z.; Shen, W.; et al. Hydrogen sulfide (H2S), a signaling molecule in plant stress responses. J. Integral Plant Biol. 2020, 63, 146–160.
- Chen, J.; Wu, F.H.; Wang, W.H.; Zheng, C.J.; Lin, G.H.; Dong, X.J.; He, J.X.; Pei, Z.M.; Zheng, H.L. Hydrogen sulphide enhances photosynthesis through promoting chloroplast biogenesis, photosynthetic enzyme expression, and thiol redox modification in Spinacia oleracea seedlings. J. Exp. Bot. 2011, 62, 4481–4493.
- Shen, J.; Zhang, J.; Zhou, M.; Zhou, H.; Cui, B.; Gotor, C.; Romero, L.C.; Fu, L.; Yang, J.; Foyer, C.H.; et al. Persulfidation-based modification of cysteine desulfhydrase and the NADPH oxidase RBOHD controls guard cell abscisic acid signaling. Plant Cell. 2020, 32, 1000–1017.
- Li, J.; Chen, S.; Wang, X.; Shi, C.; Liu, H.; Yang, J.; Shi, W.; Guo, J.; Jia, H. Hydrogen sulfide disturbs actin polymerization via S-Sulfhydration resulting in stunted root hair growth. Plant Physiol. 2018, 178, 936–949.
- Jia, H.; Chen, S.; Liu, D.; Liesche, J.; Shi, C.; Wang, J.; Ren, M.; Wang, X.; Yang, J.; Shi, W.; et al. Ethylene-induced hydrogen sulfide negatively regulates ethylene biosynthesis by persulfidation of ACO in tomato under osmotic stress. Front. Plant Sci. 2018, 9, 1517.
- Muñoz-Vargas, M.A.; González-Gordo, S.; Cañas, A.; López-Jaramillo, J.; Palma, J.M.; Corpas, F.J. Endogenous hydrogen sulfide (H2S) is up-regulated during sweet pepper (Capsicum annuum L.) fruit ripening. In vitro analysis shows that NADP-dependent isocitrate dehydrogenase (ICDH) activity is inhibited by H2S and NO. Nitric Oxide 2018, 81, 36–45.
- Muñoz-Vargas, M.A.; González-Gordo, S.; Palma, J.M.; Corpas, F.J. Inhibition of NADP-malic enzyme activity by H2S and NO in sweet pepper (Capsicum annuum L.) fruits. Physiol. Plant. 2020, 168, 278–288.
- Chen, S.; Jia, H.; Wang, X.; Shi, C.; Wang, X.; Ma, P.; Wang, J.; Ren, M.; Li, J. Hydrogen sulfide positively regulates abscisic acid signaling through persulfidation of SnRK2.6 in guard cells. Mol. Plant 2020, 13, 732–744.
- Chen, S.; Wang, X.; Jia, H.; Li, F.; Ma, Y.; Liesche, J.; Liao, M.; Ding, X.; Liu, C.; Chen, Y.; et al. Persulfidation-induced structural change in SnRK2.6 establishes intramolecular interaction between phosphorylation and persulfidation. Mol. Plant 2021, in press.
- Laureano-Marín, A.M.; Aroca, A.; Perez-Perez, M.E.; Yruela, I.; Jurado-Flores, A.; Moreno, I.; Crespo, J.L.; Romero, L.C.; Gotor, C. Abscisic acid-triggered persulfidation of cysteine protease ATG4 mediates regulation of autophagy by sulfide. Plant Cell 2020, 32, 3902–3920.
- Aroca, A.; Yruela, I.; Gotor, C.; Bassham, D.C. Persulfidation of ATG18a regulates autophagy under ER stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2023604118.
- Ma, X.; Zhang, L.; Pei, Z.; Zhang, L.; Liu, Z.; Liu, D.; Hao, X.; Jin, Z.; Pei, Y. Hydrogen sulfide promotes flowering in heading Chinese cabbage by S-sulfhydration of BraFLCs. Hortic. Res. 2021, 8, 19.
- Du, X.; Jin, Z.; Liu, Z.; Liu, D.; Zhang, L.; Ma, X.; Yang, G.; Liu, S.; Guo, Y.; Pei, Y. H2S persulfidated and increased kinase activity of MPK4 to response cold stress in Arabidopsis. Front. Mol. Biosci. 2021, 8, 635470.
- Zhou, M.; Zhang, J.; Shen, J.; Zhou, H.; Zhao, D.; Gotor, C.; Romero, L.C.; Fu, L.; Li, Z.; Yang, J.; et al. Hydrogen sulfide-linked persulfidation of ABSCISIC INSENSITIVE 4 Controls Arabidopsis ABA responses through the transactivation of mitogen-activated protein kinase kinase kinase 18. Mol. Plant 2021, 14, 921–936.
- Li, J.; Shi, C.; Wang, X.; Liu, C.; Ding, X.; Ma, P.; Wang, X.; Jia, H. Hydrogen sulfide regulates the activity of antioxidant enzymes through persulfidation and improves the resistance of tomato seedling to copper oxide nanoparticles (CuO NPs)-induced oxidative stress. Plant Physiol. Biochem. 2020, 156, 257–266.
- Corpas, F.J.; Barroso, J.B.; González-Gordo, S.; Muñoz-Vargas, M.A.; Palma, J.M. Hydrogen sulfide: A novel component in Arabidopsis peroxisomes which triggers catalase inhibition. J. Integral Plant Biol. 2019, 61, 871–883.
- Moseler, A.; Dhalleine, T.; Rouhier, N.; Couturier, J. Arabidopsis thaliana 3-mercaptopyruvate sulfurtransferases interact with and are protected by reducing systems. J. Biol. Chem. 2021, 296, 100429.
- Zhu, Y.; Liu, L.; Tan, D.; Sun, W.; Ke, Q.; Yue, X.; Bai, B. S-desulfurization: A different covalent modification mechanism from persulfidation by GSH. Free Radic. Biol. Med. 2021, 167, 54–65.
- Zaffagnini, M.; Bedhomme, M.; Lemaire, S.D.; Trost, P. The emerging roles of protein glutathionylation in chloroplasts. Plant Sci. 2012, 185–186, 86–96.
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484.
- Corpas, F.J.; Alché, J.D.; Barroso, J.B. Current overview of S-nitrosoglutathione (GSNO) in higher plants. Front. Plant Sci. 2013, 4, 126.
- Huang, J.; Willems, P.; Wei, B.; Tian, C.; Ferreira, R.B.; Bodra, N.; Martínez Gache, S.A.; Wahni, K.; Liu, K.; Vertommen, D.; et al. Mining for protein S-sulfenylation in Arabidopsis uncovers redox-sensitive sites. Proc. Natl. Acad. Sci. USA 2019, 116, 21256–21261.
- Wei, B.; Willems, P.; Huang, J.; Tian, C.; Yang, J.; Messens, J.; Van Breusegem, F. Identification of sulfenylated cysteines in Arabidopsis thaliana proteins using a disulfide-linked peptide reporter. Front. Plant Sci. 2020, 11, 777.
- Hurst, C.H.; Wright, K.M.; Turnbull, D.; Leslie, K.; Jones, S.; Hemsley, P.A. Juxta-membrane S-acylation of plant receptor-like kinases is likely fortuitous and does not necessarily impact upon function. Sci. Rep. 2019, 9, 12818.
- Hemsley, P.A. S-acylation in plants: An expanding field. Biochem. Soc. Trans. 2020, 48, 529–536.
- García, I.; Arenas-Alfonseca, L.; Moreno, I.; Gotor, C.; Romero, L.C. HCN regulates cellular processes through post-translational modification of proteins by S-cyanylation. Plant Physiol. 2019, 179, 107–123.
- Li, S.; Yu, K.; Wu, G.; Zhang, Q.; Wang, P.; Zheng, J.; Liu, Z.X.; Wang, J.; Gao, X.; Cheng, H. pCysMod: Prediction of Multiple Cysteine Modifications Based on Deep Learning Framework. Front. Cell Dev. Biol. 2021, 9, 617366.
- Wang, P.; Zhang, Q.; Li, S.; Cheng, B.; Xue, H.; Wei, Z.; Shao, T.; Liu, Z.X.; Cheng, H.; Wang, Z. iCysMod: An integrative database for protein cysteine modifications in eukaryotes. Brief. Bioinform. 2021, 7, bbaa400.
- Asada, K. Ascorbate peroxidase: A hydrogen peroxide scavenging enzyme in plants. Physiol. Plant. 1992, 85, 235–241.
- Corpas, F.J.; Trelease, R.N. Differential expression of ascorbate peroxidase and a putative molecular chaperone in the boundary membrane of differentiating cucumber seedling peroxisomes. J. Plant Physiol. 1998, 153, 332–338.
- Shigeoka, S.; Ishikawa, T.; Tamoi, M.; Miyagawa, Y.; Takeda, T.; Yabuta, Y.; Yoshimura, K. Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 2002, 53, 1305–1319.
- Kitajima, S.; Kurioka, M.; Yoshimoto, T.; Shindo, M.; Kanaori, K.; Tajima, K.; Oda, K. A cysteine residue near the propionate side chain of heme is the radical site in ascorbate peroxidase. FEBS J. 2008, 275, 470–480.
- Qu, C.; Wang, L.; Zhao, Y.; Liu, C. Molecular evolution of maize ascorbate peroxidase genes and their functional divergence. Genes 2020, 11, 1204.
- Begara-Morales, J.C.; Sánchez-Calvo, B.; Chaki, M.; Valderrama, R.; Mata-Pérez, C.; López-Jaramillo, J.; Padilla, M.N.; Carreras, A.; Corpas, F.J.; Barroso, J.B. Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. J. Exp. Bot. 2014, 65, 527–538.
- Hu, J.; Huang, X.; Chen, L.; Sun, X.; Lu, C.; Zhang, L.; Wang, Y.; Zuo, J. Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2015, 167, 1731–1746.
- Palma, J.M.; Mateos, R.M.; López-Jaramillo, J.; Rodríguez-Ruiz, M.; González-Gordo, S.; Lechuga-Sancho, A.M.; Corpas, F.J. Plant catalases as NO and H2S targets. Redox Biol. 2020, 34, 101525.
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Nitric oxide (NO) and hydrogen sulfide (H2S) modulate the NADPH-generating enzymatic system in higher plants. J. Exp. Bot. 2021, 72, 830–847.
This entry is offline, you can click here to edit this entry!
