Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Virology
Viral protein VP24 (VP24) from Ebola virus (EBOV) has been termed the minor matrix protein but its lipid-binding properties have not been investigated. Here, we found that VP24 lacks direct lipid-binding properties regardless of post-translational modification. VP24 interaction with the major matrix protein VP40 and selective plasma membrane localization were not detected, suggesting VP24 biological functions are not likely dependent on direct lipid binding.
- Ebola virus
- filovirus
- lipid binding
- matrix protein
- VP24
1. Introduction
Ebola virus (EBOV) is a lipid-enveloped filovirus with a non-segmented negative single-strand RNA genome that belongs to the order Mononegavirales. The infection of EBOV in humans can cause a highly fatal Ebola virus disease (EVD), which is characterized by acute symptoms including hemorrhagic fever, severe fatigue, emesis and multiorgan dysfunction. The high mortality of EVD was exemplified with over 11,000 deaths during the outbreak in West Africa between 2013 and 2016 [1,2] and a recent outbreak in the Democratic Republic of the Congo that claimed more than 2200 lives [3]. While an EBOV vaccine gained FDA approval late in 2019, there is still much concern regarding EBOV outbreaks as there are a lack of approved treatments for those that become infected.
The genus Ebolavirus consists of six species: Taï Forest ebolavirus (Taï Forest virus, TAFV), Reston ebolavirus (Reston virus, RESTV), Sudan ebolavirus (Sudan virus, SUDV), Bundibugyo ebolavirus (Bundibugyo virus, BDBV), Bombali ebolavirus (putatively Bombali virus, BOMV) and Zaire ebolavirus (Ebola virus, EBOV) [4,5]. Among these, EBOV is studied most intensively as it is the main circulating pathogenic ebolavirus found in infected patients in the majority of outbreaks [6].
The approximately 19 kb RNA genome of EBOV encodes at least seven proteins: the nucleoprotein (NP), the polymerase cofactor viral protein 35 (VP35), the matrix protein VP40, the transmembrane glycoprotein (GP), the transcriptional activator VP30, VP24 and the RNA-dependent RNA polymerase L [7]. The core of the Ebola virions nucleocapsid is composed of NP, VP35, VP30, VP24 and L, and encapsulates the RNA genome, while the transmembrane GP mediates virus entry and VP40 facilitates virus budding and egress at the host plasma membrane [8,9,10].
EBOV VP24 was first characterized as a minor membrane-associated protein when it was observed excluded from the nucleocapsid. The authors treated sucrose-gradient-purified Sudan viruses with 1% NP-40 lysis buffer containing various NaCl concentrations (e.g., 0.05, 0.15 and 1 M). The supernatant and pellet fractions were collected and examined by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) after centrifuging with a 30% sucrose cushion. The results showed that VP24 was not present in the pellet under medium (0.15 M) and high (1 M) salt concentrations whereas the nucleocapsid marker NP was in the pellet only, which lead to the assumption that VP24 (7.5% of virion protein) may behave as a secondary membrane associated protein like the matrix protein VP40 (37.7% of virion protein). However, VP24 was detected in both supernatant and pellet under low (0.05 M) salt conditions, the origins of which were not addressed in the previous report [11].
EBOV VP24 was also suggested to be a minor matrix protein when it was found that its cellular localization was distributed predominantly at perinuclear regions with a minor population on the plasma membrane in COS-1 cells [12]. Furthermore, the majority of VP24 protein expressed in human HEK 293T cells was found to be in the detergent phase through Trion X-114 mediated phase separation [12], where membrane associated proteins should accumulate. Lastly, VP24 was detected in the fraction collected via virus-like particle (VLP) purification method and sensitive to trypsin only in the presence of Triton X-100, indicating VP24 is incorporated into VLPs (or some lipid-encapsulated vesicle) [12]. Hence, EBOV VP24 may be a lipid-binding protein; however, the direct contribution of VP24 to VLP budding is not well understood [13].
While the lipid binding and role of VP24 in the viral matrix layer is unknown, a number of studies have shed light on the roles of VP24 in the regulation of viral transcription and replication, nucleocapsid assembly and transport, and interferon (IFN)-mediated immune signaling [14,15,16,17,18,19,20,21,22,23,24,25]. By utilizing the EBOV minigenome system, VP24 was found to inhibit viral genome transcription and replication [14,15]. VP24 is also vital for the nucleocapsid formation, condensation/maturation, transport, and thus yielding a functional virion by interacting with NP and VP35 [15,16,17,18,19,20,25]. Besides VP35, VP24 can suppress the innate immune response by preventing the nuclear accumulation of IFN-induced tyrosine-phosphorylated transcription factor STAT1 via binding host karyopherin α proteins which are nuclear localization signal receptors of activated STAT1 [21,22,23,24].
In contrast, the typical matrix proteins in filovirus such as EBOV VP40 (eVP40) and Marburg virus (MARV) VP40 (mVP40) are membrane associated peripheral proteins that bind to membrane lipids directly with high affinity. For instance, the association of eVP40 with phosphatidylserine (PS) and phosphatidylinositol-4,5-biphosphate (PI(4,5)P2) in the inner leaflet of the plasma membrane facilitates extensive eVP40 oligomerization and virus particle budding [26,27,28,29], while mVP40 tends to bind anionic membrane lipids nonspecifically through electrostatic interactions [30,31,32,33]. The expression of either eVP40 or mVP40 in mammalian cells is also sufficient to induce VLP generation [13,34,35,36].
2. Protein Expression and Purification
The protein expression constructs pET46-His-VP24, pET46-His-eVP40 and pET46-His-mVP40 were transformed into E. coli strain BL21 (DE3) pLysS. E. coli were grown at 37 °C in Luria Broth and 0.5 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG) was added into the E. coli culture when the O.D. at 600 nm was ~0.6 to induce the protein expression. The cells were collected after 12–14 h of growth at 16 °C for VP24, and at room temperature (RT) for eVP40 and mVP40. The protein purification was performed through affinity binding of the hexahistidine tag with nickel-nitrilotriacetic acid (NTA) beads (Qiagen #30230, Valencia, CA, USA) following the manufacturer′s procedures.
3. Protein–Lipid Overlay Assay
PIP strips (Echelon biosciences #P-6001, Salt Lake City, UT, USA) and membrane lipid strips (Echelon biosciences #P-6002, Salt Lake City, UT, USA) are hydrophobic membranes containing 15 different lipids (≥C16) at 100 pmol per spot along with a solvent control spot. The detailed information of each lipid species can be found on the product′s website (also see Figure 1). The lipid strips were first blocked with 3% fatty acid-free bovine serum albumin (BSA) (Fisher scientific #BP9704100, Waltham, MA, USA) in Tris-buffered saline, 0.1% Tween 20 (TBST) (20 mM Tris-HCl, pH 8.0, containing 150 mM NaCl) for 1 h at RT followed by three washes with adequate TBST buffer for 5–10 min per time. Then, 1 μg/mL of purified recombinant protein in TBST was incubated with the lipid strip overnight at 4 °C with gentle agitation. When proteins extracted from HEK293 cells were used, cells were lysed in NP-40 lysis buffer (50 mM Tris-HCl, pH 7.4, containing 150 mM NaCl, 0.1% NP-40, 1× Halt protease inhibitors) and 80 μg/ml cell lysate in TBST was incubated with lipid strips. Next, the protein solution was discarded and the lipid strip was washed three times with TBST. Afterwards, the lipid strip was incubated with monoclonal anti-polyhistidine-alkaline phosphatase antibodies (1:5000–1:10,000 dilution) in TBST for 1 h at RT. Three washes with TBST were carried out before adding alkaline phosphatase substrates (Bio-rad #1706432, Hercules, CA, USA) to detect lipid bound protein.
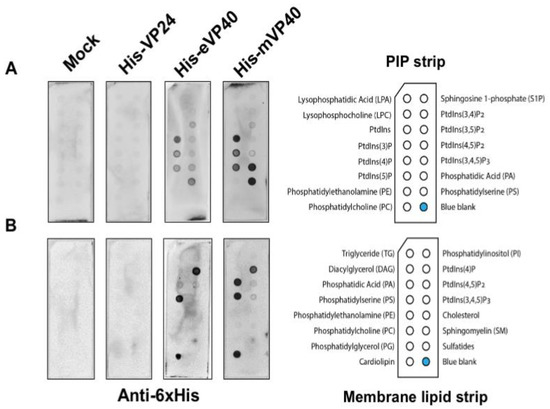
Figure 1. Ebola virus VP24 does not bind to lipids by lipid overlay assay. In this assay, 1 μg/mL of purified recombinant His-VP24, His-eVP40 and His-mVP40 proteins were incubated with PIP lipid strip (A) and membrane lipid strip (B) overnight at 4 °C. The protein purified from E. coli transformed with the empty protein expressing vector was used as the mock control. eVP40 and mVP40 bound to certain anionic lipids as expected while VP24 did not bind to any lipid tested.
4. Protein–Liposome Sendimentation Assay
To perform lipid-binding assays using a liposome sedimentation assay, liposomes were prepared using the following lipids compositions: PC:PE (70:30 mol ratio), PC:PE:PS or PA (70:20:10), and PC:PE:PIP (70:25:5). The lipid mixture was dried under a stream of nitrogen gas to completely dry the solvent (~30 min). The dried lipid films were then rehydrated in the extrusion buffer (25 mM HEPES, pH 7.5, containing 250 mM raffinose pentahydrate, 1 mM DTT) and incubated at 37 °C for at least 30 min with occasional vortexing during the incubation. The resuspended lipid solution was extruded through a 0.2-μm polycarbonate membrane 17 times by a lipid extruder to generate homogenous liposomes. The liposomes were collected after centrifuging at 50,000× g for 20 min at RT. The pelleted liposome was resuspended in the binding buffer (25 mM HEPES, pH 7.5, containing 150 mM NaCl, 0.5 mM EDTA, 1 mM DTT) and incubated with 0.5 μg of purified protein at RT for 45 min. The protein–liposome mixture was then centrifuged at 16,000× g for 30 min at RT to separate liposomes bound and free protein. Following centrifugation, the supernatant fraction was collected and the liposome pellet was resuspended in a 300–500-mL binding buffer and transferred into a new centrifuge tube subjected to another 30 min of centrifugation at 16,000× g at RT. Finally, the liposome pellet was collected and target protein was detected using Western blot in both supernatant and pellet fractions to assess the protein–liposome association.
5. EBOV VP24 Does Not Associate with Lipids
EBOV VP24 has been suggested to be a minor matrix protein and peripheral protein as the virus or cellular fraction containing the VP24 protein was sensitive to the presence of detergent [11,12]. However, to the best of our knowledge, no further direct lipid-binding studies have been examined to determine how VP24 is recruited to the virus matrix layer and plasma membrane inner leaflet. In order to determine if VP24 is a bona fide lipid-binding peripheral protein, we performed several classical lipid-binding assays. First, two types of lipid strips were used: the PIP strip containing 15 different lipids at 100 pmol per spot, which features all seven phosphoinositides (PIPs) along with a solvent control spot. Second, the membrane lipid strip possessing 15 major membrane lipids in a similar setting to the PIP strip (Figure 1; each lipid species information—see Materials and Methods section), which covers the majority of bioactive lipids.
The lipid strips were blocked with 3% fatty acid-free bovine serum albumin (BSA) and then incubated overnight with 1 μg/mL of purified recombinant 6× His-tagged VP24 (His-VP24) protein or 6× His-tagged eVP40 or mVP40 protein as positive controls at 4 °C. The lipids strips were thoroughly washed afterwards, and the lipid-bound protein was detected by monoclonal anti-polyhistidine-alkaline phosphatase antibodies. The results exhibited lack of detectable binding by VP24 to lipids on either the PIP strip or the membrane lipid strip. In contrast, eVP40 and mVP40 bound significantly to a number of anionic lipids, as previously reported, including PS [27,29,37] and phosphoinositides [28,30,32,33,38]. Lastly, no background of lipid binding was observed using purified protein from E. coli embracing an empty expression vector (Figure 1).
6. VP24 Protein Produced in Human Cells Also Does Not Associate with Lipids
Next, we hypothesized that VP24 lipid-binding may be more dependent on mammalian cell expression due to potential post-translational modifications (PTMs) that may alter VP24–lipid interactions. For instance, VP24 is known to undergo SUMOylation that covalently attaches the small ubiquitin-like modifier (SUMO) to the target protein [39]. Thus, HEK293 cells were transfected with plasmids of HA-tagged VP24 (HA-VP24) or enhanced green fluorescent protein (EGFP)-tagged lactadherin C2 domain (LactC2) (LactC2-EGFP) which is a well-established PS binding protein [40]. First, the protein expression of HA-VP24 and LactC2-EGFP from HEK293 cells was examined (Figure 2A) demonstrating robust soluble protein expression. Next, the membrane lipid strips described above were incubated overnight at 4 °C with 80 μg/mL of cell lysate of HEK 293 cells, HEK 293 cells expressing HA-VP24 or HEK 293 cells expressing LactC2-EGFP. The primary anti-HA and anti-GFP antibodies along with the correlated secondary antibodies were applied to detect lipid-bound HA-VP24 and LactC2-EGFP, respectively. Similar to the mock control, no lipid-bound HA-VP24 was observed, while LactC2-EGFP bound to PS specifically as expected (Figure 2B), indicating that mammalian expression of VP24 had little impact on its lipid-binding ability.
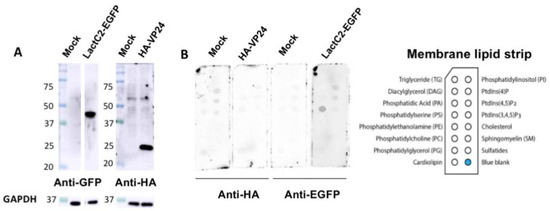
Figure 2. VP24 expressed in HEK293 cells did not bind to lipids. (A). The expression of LactC2-EGFP and HA-VP24 were confirmed by Western blot. (B). The 80 μg/mL of cell lysate expressing indicated proteins was incubated with membrane lipid strips overnight at 4 °C. The cell lysate of HEK 293 cells with mock transfection was used as a control. HA-VP24 did not bind to any lipids while LactC2-EGFP bound to PS as expected.
References
- Rojas, M.; Monsalve, D.M.; Pacheco, Y.; Acosta-Ampudia, Y.; Ramírez-Santana, C.; Ansari, A.A.; Gershwin, M.E.; Anaya, J.-M. Ebola virus disease: An emerging and re-emerging viral threat. J. Autoimmun. 2020, 106, 102375. [Google Scholar] [CrossRef] [PubMed]
- Fanunza, E.; Frau, A.; Sgarbanti, M.; Orsatti, R.; Corona, A.; Tramontano, E. Development and Validation of a Novel Dual Luciferase Reporter Gene Assay to Quantify Ebola Virus VP24 Inhibition of IFN Signaling. Viruses 2018, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- WHO. Ebola Virus Disease—Democratic Republic of the Congo. Available online: https://www.who.int/csr/don/04-June-2020-ebola-drc/en/ (accessed on 6 July 2020).
- Matson, M.J.; Chertow, D.S.; Munster, V.J. Delayed recognition of Ebola virus disease is associated with longer and larger outbreaks. Emerg. Microbes Infect. 2020, 9, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, T.; Anthony, S.J.; Gbakima, A.; Bird, B.H.; Bangura, J.; Tremeau-Bravard, A.; Belaganahalli, M.N.; Wells, H.L.; Dhanota, J.; Liang, E.; et al. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat. Microbiol. 2018, 3, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Adachi, T.; Adhikari, N.K.J.; Arribas, J.R.; Bah, I.E.; Bausch, D.G.; Bhadelia, N.; Borchert, M.; Brantsæter, A.B.; Brett-Major, D.M.; et al. New filovirus disease classification and nomenclature. Nat. Rev. Genet. 2019, 17, 261–263. [Google Scholar] [CrossRef]
- Baseler, L.; Chertow, D.S.; Johnson, K.M.; Feldmann, H.; Morens, D.M. The Pathogenesis of Ebola Virus Disease. Annu. Rev. Pathol. Mech. Dis. 2017, 12, 387–418. [Google Scholar] [CrossRef]
- Martin, B.; Reynard, O.; Volchkov, V.; Decroly, E. Filovirus proteins for antiviral drug discovery: Structure/function of proteins involved in assembly and budding. Antivir. Res. 2018, 150, 183–192. [Google Scholar] [CrossRef]
- Stahelin, R.V. Membrane binding and bending in Ebola VP40 assembly and egress. Front. Microbiol. 2014, 5, 300. [Google Scholar] [CrossRef]
- Hoenen, T.; Groseth, A.; Feldmann, H. Therapeutic strategies to target the Ebola virus life cycle. Nat. Rev. Genet. 2019, 17, 593–606. [Google Scholar] [CrossRef]
- Elliott, L.H.; Kiley, M.P.; McCormick, J.B. Descriptive analysis of Ebola virus proteins. Virology 1985, 147, 169–176. [Google Scholar] [CrossRef]
- Han, Z.; Boshra, H.; Sunyer, J.O.; Zwiers, S.H.; Paragas, J.; Harty, R.N. Biochemical and Functional Characterization of the Ebola Virus VP24 Protein: Implications for a Role in Virus Assembly and Budding. J. Virol. 2003, 77, 1793–1800. [Google Scholar] [CrossRef] [PubMed]
- Licata, J.M.; Johnson, R.F.; Han, Z.; Harty, R.N. Contribution of Ebola Virus Glycoprotein, Nucleoprotein, and VP24 to Budding of VP40 Virus-Like Particles. J. Virol. 2004, 78, 7344–7351. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Noda, T.; Halfmann, P.; Jasenosky, L.; Kawaoka, Y. Ebola Virus (EBOV) VP24 Inhibits Transcription and Replication of the EBOV Genome. J. Infect. Dis. 2007, 196, S284–S290. [Google Scholar] [CrossRef] [PubMed]
- Mateo, M.; Carbonnelle, C.; Martínez, M.J.; Reynard, O.; Page, A.; Volchkova, V.A.; Volchkov, V.E. Knockdown of Ebola Virus VP24 Impairs Viral Nucleocapsid Assembly and Prevents Virus Replication. J. Infect. Dis. 2011, 204, S892–S896. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, L.; Sun, Y.; Nabel, G.J. The Assembly of Ebola Virus Nucleocapsid Requires Virion-Associated Proteins 35 and 24 and Posttranslational Modification of Nucleoprotein. Mol. Cell 2002, 10, 307–316. [Google Scholar] [CrossRef]
- Noda, T.; Halfmann, P.; Sagara, H.; Kawaoka, Y. Regions in Ebola Virus VP24 That Are Important for Nucleocapsid Formation. J. Infect. Dis. 2007, 196, S247–S250. [Google Scholar] [CrossRef]
- Watt, A.; Moukambi, F.; Banadyga, L.; Groseth, A.; Callison, J.; Herwig, A.; Ebihara, H.; Feldmann, H.; Hoenen, T. A Novel Life Cycle Modeling System for Ebola Virus Shows a Genome Length-Dependent Role of VP24 in Virus Infectivity. J. Virol. 2014, 88, 10511–10524. [Google Scholar] [CrossRef]
- Banadyga, L.; Hoenen, T.; Ambroggio, X.; Dunham, E.; Groseth, A.; Ebihara, H. Ebola virus VP24 interacts with NP to facilitate nucleocapsid assembly and genome packaging. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Takamatsu, Y.; Kolesnikova, L.; Becker, S. Ebola Virus Proteins NP, VP35, and VP24 are Essential and Sufficient to Mediate Nucleocapsid Transport. Proc. Natl. Acad. Sci. USA 2018, 115, 1075–1080. [Google Scholar] [CrossRef]
- Reid, S.P.; Leung, L.W.; Hartman, A.L.; Martinez, O.; Shaw, M.L.; Carbonnelle, C.; Volchkov, V.E.; Nichol, S.T.; Basler, C.F. Ebola Virus VP24 Binds Karyopherin α1 and Blocks STAT1 Nuclear Accumulation. J. Virol. 2006, 80, 5156–5167. [Google Scholar] [CrossRef]
- Reid, S.P.; Valmas, C.; Martinez, O.; Sanchez, F.M.; Basler, C.F. Ebola Virus VP24 Proteins Inhibit the Interaction of NPI-1 Subfamily Karyopherin α Proteins with Activated STAT1. J. Virol. 2007, 81, 13469–13477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.P.P.; Bornholdt, Z.A.; Liu, T.; Abelson, D.M.; Lee, D.E.; Li, S.; Woods, V.L.; Saphire, E.O. The Ebola Virus Interferon Antagonist VP24 Directly Binds STAT1 and Has a Novel, Pyramidal Fold. PLoS Pathog. 2012, 8, e1002550. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Edwards, M.R.; Borek, D.; Feagins, A.R.; Mittal, A.; Alinger, J.B.; Berry, K.N.; Yen, B.; Hamilton, J.R.; Brett, T.J.; et al. Ebola Virus VP24 Targets a Unique NLS Binding Site on Karyopherin Alpha 5 to Selectively Compete with Nuclear Import of Phosphorylated STAT1. Cell Host Microb. 2014, 16, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, Y.; Kolesnikova, L.; Schauflinger, M.; Noda, T.; Becker, S. The Integrity of the YxxL Motif of Ebola Virus VP24 Is Important for the Transport of Nucleocapsid-Like Structures and for the Regulation of Viral RNA Synthesis. J. Virol. 2020, 94, 2170–2179. [Google Scholar] [CrossRef]
- Bornholdt, Z.A.; Noda, T.; Abelson, D.M.; Halfmann, P.; Wood, M.R.; Kawaoka, Y.; Saphire, E.O. Structural Rearrangement of Ebola Virus VP40 Begets Multiple Functions in the Virus Life Cycle. Cell 2013, 154, 763–774. [Google Scholar] [CrossRef]
- Adu-Gyamfi, E.; Johnson, K.A.; Fraser, M.E.; Scott, J.L.; Soni, S.P.; Jones, K.R.; Digman, M.A.; Gratton, E.; Tessier, C.R.; Stahelin, R.V. Host Cell Plasma Membrane Phosphatidylserine Regulates the Assembly and Budding of Ebola Virus. J. Virol. 2015, 89, 9440–9453. [Google Scholar] [CrossRef]
- Johnson, K.A.; Taghon, G.J.F.; Scott, J.L.; Stahelin, R.V. The Ebola Virus matrix protein, VP40, requires phosphatidylinositol 4,5-bisphosphate (PI (4,5) P2) for extensive oligomerization at the plasma membrane and viral egress. Sci. Rep. 2016, 6, srep19125. [Google Scholar] [CrossRef]
- Del Vecchio, K.; Frick, C.T.; Gc, J.B.; Oda, S.-I.; Gerstman, B.S.; Saphire, E.O.; Chapagain, P.P.; Stahelin, R.V. A cationic, C-terminal patch and structural rearrangements in Ebola virus matrix VP40 protein control its interactions with phosphatidylserine. J. Boil. Chem. 2018, 293, 3335–3349. [Google Scholar] [CrossRef]
- Wijesinghe, K.J.; Stahelin, R.V. Investigation of the Lipid Binding Properties of the Marburg Virus Matrix Protein VP40. J. Virol. 2015, 90, 3074–3085. [Google Scholar] [CrossRef]
- Oda, S.-I.; Noda, T.; Wijesinghe, K.J.; Halfmann, P.; Bornholdt, Z.A.; Abelson, D.M.; Armbrust, T.; Stahelin, R.V.; Kawaoka, Y.; Saphire, E.O. Crystal Structure of Marburg Virus VP40 Reveals a Broad, Basic Patch for Matrix Assembly and a Requirement of the N-Terminal Domain for Immunosuppression. J. Virol. 2015, 90, 1839–1848. [Google Scholar] [CrossRef]
- Wijesinghe, K.J.; Urata, S.; Bhattarai, N.; Kooijman, E.E.; Gerstman, B.S.; Chapagain, P.P.; Li, S.; Stahelin, R.V. Detection of lipid-induced structural changes of the Marburg virus matrix protein VP40 using hydrogen/deuterium exchange-mass spectrometry. J. Boil. Chem. 2017, 292, 6108–6122. [Google Scholar] [CrossRef] [PubMed]
- Wijesinghe, K.J.; McVeigh, L.; Husby, M.L.; Bhattarai, N.; Ma, J.; Gerstman, B.S.; Chapagain, P.P.; Stahelin, R.V. Mutation of Hydrophobic Residues in the C-Terminal Domain of the Marburg Virus Matrix Protein VP40 Disrupts Trafficking to the Plasma Membrane. Viruses 2020, 12, 482. [Google Scholar] [CrossRef] [PubMed]
- Jasenosky, L.D.; Neumann, G.; Lukashevich, I.; Kawaoka, Y. Ebola Virus VP40-Induced Particle Formation and Association with the Lipid Bilayer. J. Virol. 2001, 75, 5205–5214. [Google Scholar] [CrossRef]
- Kolesnikova, L.; Berghöfer, B.; Bamberg, S.; Becker, S. Multivesicular Bodies as a Platform for Formation of the Marburg Virus Envelope. J. Virol. 2004, 78, 12277–12287. [Google Scholar] [CrossRef]
- Swenson, D.L.; Warfield, K.L.; Kuehl, K.; Larsen, T.; Hevey, M.; Schmaljohn, A.; Bavari, S.; Aman, M.J. Generation of Marburg virus-like particles by co-expression of glycoprotein and matrix protein. FEMS Immunol. Med. Microbiol. 2004, 40, 27–31. [Google Scholar] [CrossRef]
This entry is adapted from the peer-reviewed paper 10.3390/v12080869
This entry is offline, you can click here to edit this entry!
