Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Genetics & Heredity
The purpose of this study was to classify breast cancer subtypes by using deep neural networks based on multi-omics data from TCGA. The classification results show that, by using the proposed model, integrating multi-omics datasets can improve the performance as compared to using single omics data for classifying breast cancer subtypes.
- omics data integration
- breast cancer subtype
- deep neural networks
1. Introduction
Breast cancer is the most common cancer and the main cause of cancer deaths besides lung cancer in women [1]. The number of breast cancer patients is increasing year by year, and the proportion of women under 40 who have breast cancer has already reached 6.6% [2]. In 2018, there were more than 2 million new breast cancer cases worldwide [3]. At the same time, as a highly heterogeneous disease, breast cancer is composed of different biological subtypes, with different clinical, pathological, and molecular characteristics, as well as prognostic and therapeutic significance [4]. Therefore, the study of breast cancer subtypes is of great significance for precision medicine and prognosis prediction of breast cancer [5]. By understanding the molecular subtypes of breast cancer, doctors can better decide which treatment is suitable for each patient, thus saving money for the whole medical system and avoiding the side effects of unnecessary treatment [6].
The current research on breast cancer subtypes focuses mainly on the molecular typing. In 1999, molecular typing of cancer was first proposed by the National Cancer Institute (NCI) [6]. In 2000, Perou et al. first proposed the molecular typing of breast cancer and concluded that breast cancer is divided into four subtypes, namely luminal subtype, basal-like subtype, human epidermal growth subtype and normal breast-like subtype [7]. Sorlie et al. further subdivided luminal subtype into luminal A and luminal B [8]. Waks et al. classified breast cancer into three major subtypes based on the presence or absence of molecular markers for estrogen receptor (ER) and progesterone receptor (PR) and human epidermal growth factor 2 (HER2), namely ER+/PR+/HER2- (luminal A), HER2+, and triple-negative breast cancer (TNBC), which have a negative indicator in all three standard molecular markers [9]. HER2+ subtype can be further divided into two subtypes: ER+/PR+/HER2+ (luminal B) and ER-/PR-/HER2+. Tao et al. classified breast cancer into five subtypes according to immunohistochemistry (IHC) markers, including ER, PR, and HER2 [6]. These subtypes are luminal A, luminal B, HER2(+), TNBC, and unclear subtype.
In this article, breast cancer was divided into five subtypes, namely luminal A, luminal B, HER2(+), TNBC, and unclear subtype, the same classification as in a published article [10]. The detailed definition of each subtype is shown in Table 1. Luminal A is the most common breast cancer subtype, accounting for as many as 60% of all breast cancers [11]. This subtype has the highest prognosis among several breast cancer subtypes, and its 5-year local recurrence rate is much lower than other breast cancer subtypes [12]. Most patients with luminal B are elderly patients. They are similar to luminal A in that they are also sensitive to endocrine therapy. Hormone expression in patients with luminal B is reported to be lower than that of luminal A, whereas the expression and histological grade of proliferation markers are higher than those of luminal A [13]. HER2-positive breast cancer patients account for about 25%, and the prognosis is poor. Most patients with advanced HER2-positive breast cancer are most likely to metastasize to the axillary lymph nodes. In the treatment, endocrine therapy has almost no effect on it. The TNBC subtype has ER negative, PR negative, and HER2-negative [6]. Compared with other breast cancer subtypes, TNBC has rapid deterioration and metastasis. Because its three receptors are negative, targeted therapy cannot be used during the treatment of this subtype, and its prognosis is poor. Unclear subtype refers to patient samples that lack information on each of the three IHC markers.
Table 1. The detailed definition of breast cancer subtypes.
| Breast Cancer Subtypes | IHC Markers |
|---|---|
| luminal A | ER/PR+, Her2− |
| luminal B | ER/PR+, Her2+ |
| HER2(+) | ER/PR−, Her2+ |
| TNBC | ER/PR−, Her2− |
| unclear | lacking |
With the explosive growth of massive biological data, the transformation of traditional biological statistical methods to computer-aided methods makes machine learning become an important part of predicting cancer prognosis [14]. If all the features in these samples are used to classify and regress, it will lead to overfitting. Feature selection or reduction, which attempts to find the subset of features that gives the model the best performance, is one of the solutions [15]. Utilizing the feature selection method can remove the obviously irrelevant and redundant gene features and improve the performance of the model. Furthermore, fewer features usually mean better interpretability and higher training speed in deep neural networks.
At present, the commonly used feature selection methods are mainly divided into the following three types: filter, wrapper, and embedded [16]. These categories are mainly based on the combination of search process of feature selection and construction process of classification models [17]. The filter methods are independent of the classifier and only rely on the intrinsic attributes of the data to select relevant features [18]. In the wrapper methods, the classification score of the features by the classifier is measured during the selection process, and the feature selection step depends on the classifier [19]. In other words, the wrapper feature selection method is to select the most fruitful feature subset for the given classifier. In the embedded methods, the step of selecting the optimal feature subset is embedded in the construction of the classifier, and the selection process can be regarded as the combined space of feature subsets and hypotheses [16]. They are completed in the same optimization process. It means that feature selection is automatically carried out during learner training. In general, when comparing complex wrappers and embedded methods to the filtering methods, the computational complexity of the former two methods is always higher, and the performance is not as good as the simple filtering method [20].
With the continuous development and improvement of high-throughput technology, there are increasingly more types of omics data obtained through high-throughput technology. Based on these omics data, there have been many studies on the classification of breast cancer subtypes. Brian D. Lehmann et al. used gene expression data to perform cluster analysis to determine the subtypes of triple-negative breast cancer [21]. Sorlie et al. achieved the classification of breast cancer subtypes by constructing a gene expression pattern based on hierarchical clustering [22]. Each type of omics data itself usually provides a list of differences associated with the disease [23]. However, the analysis of one type of omics data is limited to correlation, mainly reflecting the reaction process rather than the causal process. Multi-omics data are expected to improve the characterization of cross-molecular biological processes, and can provide more comprehensive insights into the biological systems being studied [24]. The use of multi-omics data for cancer classification has been recently suggested [25]. Multi-omics data have been used to solve different problems such as precision oncology [26], driver gene identification [27], regulatory genomics [28], and drug response prediction [29]. Artificial intelligence in cancer science includes not only classification but also diagnostics [30] as well as prediction of clinical features or identification of interactions. Most importantly, it includes research integrating multi-omics data type [31]. A further example, such as in [32], could be likewise included, as well as the one in [33]. However, there are currently few studies on the classification of breast cancer subtypes based on multi-omics data. Tao et al. used multiple kernel learning (MKL) based on multi-omics data to classify breast cancer subtypes [6]. MKL is a method widely used in multi-omics data fusion, which can improve the classification performance of original (Support Vector Machine) SVM [34]. For the classification of breast cancer subtypes, various kernels are generated and normalized using different omics data. Subsequently, after training the MKL model based on these kernels, other multi-omics data can be used to predict based on the trained model.
In this study, DeepMO, a model using deep neural networks based on multi-omics data, was employed to classify breast cancer subtypes. DeepMO contains a type-specific encoding subnetwork to learn the features of each omics type and combines features of each omics type, and a classification subnetwork is used to classify different breast cancer subtypes. In this study, the input of DeepMO contains mRNA data, DNA methylation data, and copy number variation (CNV) data, and the output of DeepMO is the predicted molecular subtypes of breast cancer. The workflow of DeepMO is illustrated in Figure 1. We compared the performances of binary classification based on multi-omics data and single omics data. Moreover, the performances of binary classification using DeepMO and MKL [6] were also compared. Then, we compared the performances of multi-classification based on multi-omics data and single omics data. Additionally, the performance of DeepMO on multi-classification was compared with some state-of-the-art data integration methods. Furthermore, we analyzed the effect of feature selection, and validated its role in classification using deep neural networks based on multi-omics data. Finally, we also analyzed the enriched gene ontology (GO) terms and biological pathways of these significant genes discovered during the feature selection process.
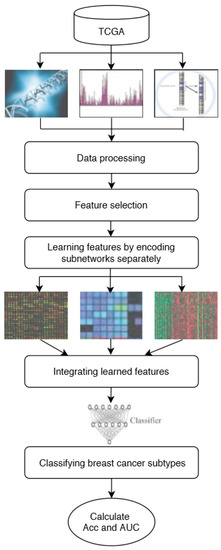
Figure 1. The workflow of DeepMO.
2. The Performance of Multi-Classification
To better evaluate the performance of the proposed model, we used the model to predict breast cancer subtypes based on multi-classification. First, we compared the accuracy of multi-classification between DeepMO using single omics data and multiple omics data. The mean accuracy of 5-fold cross-validation on all subtypes of breast cancer by using single omics data and combining three omics data is shown in Figure 4. From the figure, we can conclude that DeepMO was superior to single omics data on multi-classification.
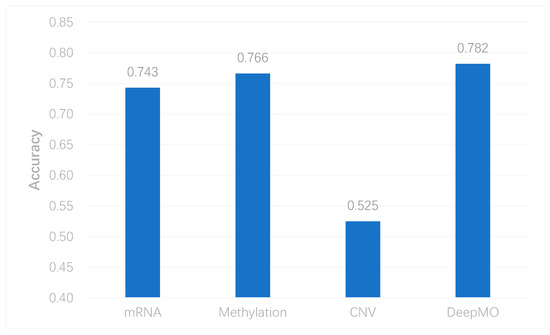
Figure 4. The accuracy by using single omics data and multi-omics data.
Due to the imbalance of class, which had an effect on the neural network, we took some measures to cope with it. In this study, we used both the undersampling and oversampling methods to reduce the effect of imbalanced data. We selected samples by the weight of each subtype and the weight was reciprocal to the number of each subtype. Therefore, the subtype with smaller samples had more probability to be selected. This may make our model applicable to imbalanced data. To evaluate this method, we removed all samples of one subtype each time, and performed multi-classification again. The results are shown in Figure 5. The accuracy of DeepMO is relatively stable and it indicates that imbalanced data have no dramatic impact on our model.
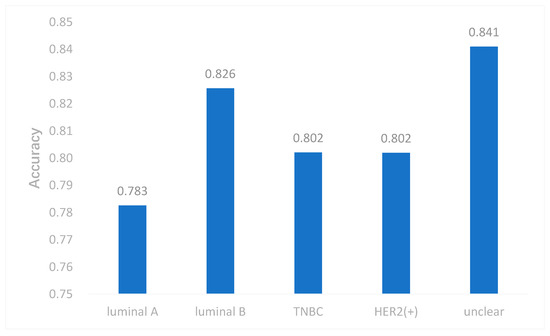
Figure 5. The accuracy of DeepMO when removing one subtype each time.
To further evaluate the performance of DeepMO on multi-classification, we selected some state-of-the-art methods of omics data integration, including the logistic regression model/multinational model with Elastic Net (EN) regularization [41] and Random Forest (RF) [42] in the concatenation and ensemble frameworks apart from MKL [6]. They were denoted as ConcateEN, ConcateRF, EnsembleEN, and EnsembleRF, respectively. The accuracy of multi-classification using different methods is shown in Figure 6. We can observe that DeepMO outperformed other methods in multi-classification.
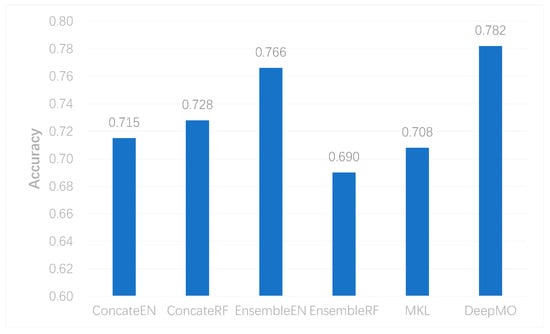
Figure 6. The accuracy of multi-classification using different methods.
3. Analysis of the Effect of Feature Selection
We utilized the chi-squared test to select the top 5000 important features before integrating multi-omics data using deep neural networks for reducing features and increasing training speed. The top 5000 features of three omics data types for multi-classification are shown in the Supplementary Materials. Furthermore, we assumed that the feature selection algorithms can further improve the deep neural network model. To better evaluate the ability to classify each subtype, we compared the results with and without feature selection both on binary and multi-classification. The results of multi-omics data integration using deep neural networks with and without feature selection are shown in Figure 7 and Figure 8. From Figure 7, we can conclude that using feature selection can improve the accuracy on binary classification. From Figure 8, it is clear that when using feature selection, the AUC on binary classification can also be improved.
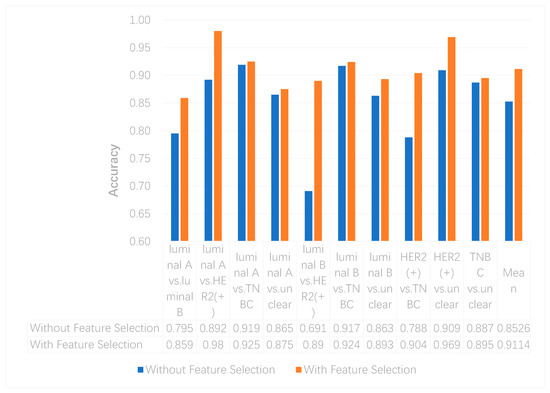
Figure 7. The accuracy of multi-omics data integration using deep neural networks with and without feature selection.
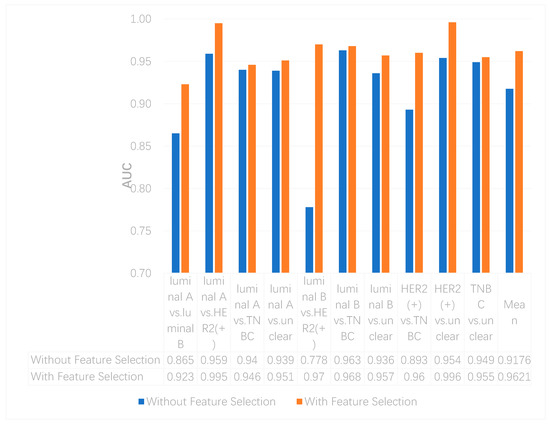
Figure 8. The AUCs of multi-omics data integration using deep neural networks with and without feature selection.
Additionally, our experiments indicated that feature selection can improve accuracy on multi-classification. The accuracy without and with feature selection on multi-classification is 0.771 and 0.782, respectively.
References
- Callahan, R.; Hurvitz, S.A. Human epidermal growth factor receptor-2-positive breast cancer: Current management of early, advanced, and recurrent disease. Curr. Opin. Obstet. Gynecol. 2011, 23, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Assi, H.A.; Khoury, K.E.; Dbouk, H.; Khalil, L.E.; Mouhieddine, T.H.; Saghir, M.Z. Epidemiology and prognosis of breast cancer in young women. J. Thorac. Dis. 2013, 5, S2–S8. [Google Scholar] [PubMed]
- Maurer Foundation Breast Health Education. Breast Cancer Statistics. Available online: https://www.maurerfoundation.org/about-breast-cancer-breast-health/breast-cancer-statistics/ (accessed on 24 May 2020).
- Reis-Filho, J.S.; Pusztai, L. Gene expression profiling in breast cancer: Classification, prognostication, and prediction. Lancet 2011, 378, 1812–1823. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Tao, M.; Song, T.; Du, W.; Han, S.; Zuo, C.; Li, Y.; Wang, Y.; Yang, Z. Classifying Breast Cancer Subtypes Using Multiple Kernel Learning Based on Omics Data. Genes 2019, 10, 200. [Google Scholar] [CrossRef]
- Perou, C.M.; Serlie, T.; Eisen, M.B.; Matt, V.D.R.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A. Molecular portraits of human breast tumors. Nature 2012, 490, 747–752. [Google Scholar]
- Sørlie, T.; Tibshirani, R.; Parker, J.; Hastie, T.; Marron, J.S.; Nobel, A.; Deng, S.; Johnsen, H.; Pesich, R.; Geisler, S.; et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Nat. Acad. Sci. USA 2003, 100, 8418–8423. [Google Scholar]
- Yersal, Ö.; Barutca, S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World J. Clin. Oncol. 2014, 5, 412–424. [Google Scholar] [CrossRef]
- Mylan. Understanding the Differences between Breast Cancer Subtypes. Available online: https://www.mylan.com/en/news/feature-stories/understanding-breast-cancer-subtypes-october-2019 (accessed on 24 May 2020).
- Ebili, H.; Oluwasola, A.; Olopade, O. Molecular subtypes of breast cancer. In Personalized Management of Breast Cancer; Holloway, T.L., Jatoi, I., Eds.; Future Medicine Ltd.: London, UK, 2014; pp. 20–33. [Google Scholar] [CrossRef]
- Nguyen, P.L.; Taghian, A.G.; Katz, M.S.; Niemierko, A.; Raad, R.F.A.; Boon, W.L.; Bellon, J.R.; Wong, J.S.; Smith, B.L.; Harris, J.R. Breast Cancer Subtype Approximated by Estrogen Receptor, Progesterone Receptor, and HER-2 Is Associated with Local and Distant Recurrence After Breast-Conserving Therapy. J. Clin. Oncol. 2008, 26, 2373–2378. [Google Scholar] [CrossRef]
- Ades, F.; Zardavas, D.; Bozovic-Spasojevic, I.; Pugliano, L.; Fumagalli, D.; De Azambuja, E.; Viale, G.; Sotiriou, C.; Piccart, M. Luminal B Breast Cancer: Molecular Characterization, Clinical Management, and Future Perspectives. J. Clin. Oncol. 2014, 32, 2794–2803. [Google Scholar] [CrossRef]
- Ma, B.; Meng, F.; Yan, G.; Yan, H.; Chai, B.; Song, F. Diagnostic classification of cancers using extreme gradient boosting algorithm and multi-omics data. Comput. Boil. Med. 2020, 121, 103761. [Google Scholar] [CrossRef] [PubMed]
- Ge, R.; Zhou, M.; Luo, Y.; Meng, Q.; Mai, G.; Ma, D.; Wang, G.; Zhou, F. McTwo: A two-step feature selection algorithm based on maximal information coefficient. BMC Bioinform. 2016, 17, 142. [Google Scholar] [CrossRef] [PubMed]
- Guyon, I.; André, E. An introduction to variable and feature selection. J. Mach Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Cao, Z.; Wang, Y.; Sun, Y.; Du, W.; Liang, Y. A novel filter feature selection method for paired microarray expression data analysis. Int. J. Data Min. Bioinform. 2015, 12, 363. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Cao, Z.; Song, T.; Li, Y.; Liang, Y. A feature selection method based on multiple kernel learning with expression profiles of different types. BioData Min. 2017, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, S.; Weber, R. A wrapper method for feature selection using Support Vector Machines. Inf. Sci. 2009, 179, 2208–2217. [Google Scholar] [CrossRef]
- Haury, A.-C.; Gestraud, P.; Vert, J.-P. The Influence of Feature Selection Methods on Accuracy, Stability and Interpretability of Molecular Signatures. PLoS ONE 2011, 6, e28210. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Nat. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L. Deep Learning–Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res. 2017, 24, 1248–1259. [Google Scholar] [CrossRef]
- Argelaguet, R.; Velten, B.; Arnol, D.; Dietrich, S.; Zenz, T.; Marioni, J.C.; Buettner, F.; Huber, W.; Stegle, O. Multi-Omics Factor Analysis—A framework for unsupervised integration of multi-omics data sets. Mol. Syst. Boil. 2018, 14, e8124. [Google Scholar] [CrossRef] [PubMed]
- Bavafaye, H.; Elham, M.K.; Britt, E.L.; Søren, B. Hierarchical Classification of Cancers of Unknown Primary Using Multi-Omics Data. Cancer Inform. 2019, 18, 1176935119872163. [Google Scholar]
- Ding, M.; Chen, L.; Cooper, G.F.; Young, J.D.; Lu, X. Precision Oncology beyond Targeted Therapy: Combining Omics Data with Machine Learning Matches the Majority of Cancer Cells to Effective Therapeutics. Mol. Cancer Res. 2017, 16, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Dimitrakopoulos, C.; Hindupur, S.K.; Häfliger, L.; Behr, J.; Montazeri, H.; Hall, M.N.; Beerenwinkel, N. Network-based integration of multi-omics data for prioritizing cancer genes. Bioinformatics 2018, 34, 2441–2448. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ge, B.; Casale, F.P.; Vasquez, L.; Kwan, T.; Garrido-Martín, D.; Watt, S.; Yan, Y.; Kundu, K.; Ecker, S.; et al. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell 2016, 167, 1398–1414.e24. [Google Scholar] [CrossRef] [PubMed]
- Sharifi-Noghabi, H.; Zolotareva, O.I.; Collins, C.; Ester, M. MOLI: Multi-omics late integration with deep neural networks for drug response prediction. Bioinformatics 2019, 35, i501–i509. [Google Scholar] [CrossRef] [PubMed]
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Back, T.; Chesus, M.; Corrado, G.C.; Darzi, A.; et al. International evaluation of an AI system for breast cancer screening. Nature 2020, 577, 89–94. [Google Scholar] [CrossRef]
- Jeanquartier, F.; Jean-Quartier, C.; Kotlyar, M.; Tokar, T.; Hauschild, A.-C.; Jurisica, I.; Holzinger, A. Machine Learning for In Silico Modeling of Tumor Growth. In Computer Vision; Springer Science and Business Media LLC: Berlin, Germany, 2016; Volume 9605, pp. 415–434. [Google Scholar]
- Yan, R.; Ren, F.; Rao, X.; Shi, B.; Xiang, T.; Zhang, L.; Liu, Y.; Liang, J.; Zheng, C.; Zhang, F. Integration of Multimodal Data for Breast Cancer Classification Using a Hybrid Deep Learning Method. In Proceedings of the Intelligent Tutoring Systems; Springer Science and Business Media LLC: Berlin, Germany, 2019; pp. 460–469. [Google Scholar]
- Guo, Y.; Shang, X.; Li, Z. Identification of cancer subtypes by integrating multiple types of transcriptomics data with deep learning in breast cancer. Neurocomputing 2018, 324, 20–30. [Google Scholar] [CrossRef]
- Qi, J.; Xun, L.; Xu, R. A Multiple Kernel Learning Model Based on p-Norm. Comput. Intell. Neurosci. 2018, 2018, 1–7. [Google Scholar] [CrossRef]
This entry is adapted from the peer-reviewed paper 10.3390/genes11080888
This entry is offline, you can click here to edit this entry!
