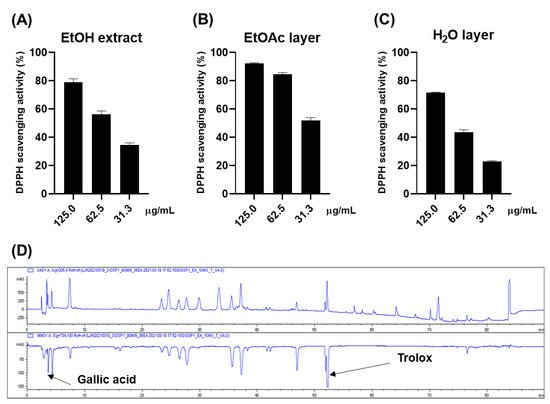
Diospyros kaki (persimmon) leaves have long been utilized as traditional medicine for the treatment of ischemic stroke, angina, and hypertension and as a healthy beverage and cosmetic for anti-aging. This study aimed to isolate as many compounds as possible from an ethanol extract of the persimmon leaves to identify the biologically active compounds. The antioxidative effect of the ethyl acetate layer from the ethanol extract of the persimmon leaves was demonstrated by 2,2-diphenyl-1-picrylhydrazyl (DPPH) assay and online high-performance liquid chromatography-2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (HPLC-ABTS) analysis.
- Diospyros kaki
- persimmon leaves
- flavonoid
- antioxidant
1. Introduction
2. Antioxidative Effect of the Persimmon Leaves
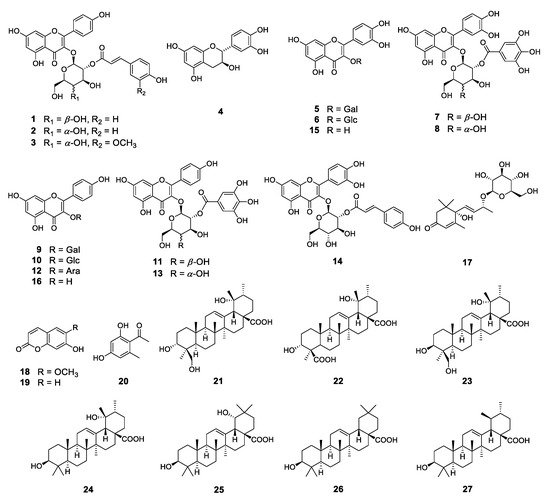
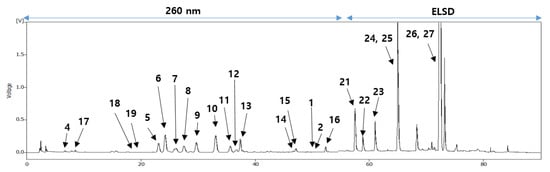
3. Phytochemical Investigation
| Number of Carbon |
1 | 3 | ||
|---|---|---|---|---|
| δH Multi (J in Hz) | δC | δH Multi (J in Hz) | δC | |
| 2 | 158.1 | 158.6 | ||
| 134.9 | 134.8 | |||
| 4 | 179.2 | 179.0 | ||
| 5 | 163.1 | 163.0 | ||
| 6 | 6.16 d (1.5) | 101.2 | 6.11 d (2.0) | 100.6 |
| 7 | 167.7 | 168.3 | ||
| 8 | 6.34 s | 95.1 | 6.29 d (2.0) | 95.2 |
| 9 | 158.5 | 158.1 | ||
| 10 | 105.3 | 105.2 | ||
| 1′ | 122.7 | 122.8 | ||
| 2′,6′ | 8.00 d (8.5) | 132.1 | 7.98 d (9.0) | 132.1 |
| 3′,5′ | 6.87 d (8.5) | 116.3 | 6.88 d (9.0) | 116.2 |
| 4′ | 161.6 | 161.6 | ||
| 1′′ | 5.57 d (8.0) | 100.4 | 5.64 d (8.0) | 100.7 |
| 2′′ | 5.36 dd (10.0, 8.0) | 74.3 | 5.03 dd (9.0, 8.0) | 75.8 |
| 3′′ | 3.75 dd (10.5, 3.5) | 73.4 | 3.64 t (9.0) | 76.3 |
| 4′′ | 3.89 d (3.5) | 70.5 | 3.41 t (10.0) | 71.5 |
| 5′′ | 3.55 t (6.0) | 77.4 | 3.29 m | 78.8 |
| 6′′ | 3.67 m | 62.0 | 3.78 dd (12.0, 2.0) | 62.5 |
| 3.61 m | ||||
| 1′′′ | 127.2 | 127.8 | ||
| 2′′′ | 7.45 d (8.5) | 131.2 | 7.18 d (1.5) | 111.7 |
| 3′′′ | 6.81 d (8.5) | 116.8 | 149.4 | |
| 4′′′ | 161.3 | 150.7 | ||
| 5′′′ | 6.81 d (8.5) | 116.8 | 6.81 d (8.5) | 116.5 |
| 6′′′ | 7.45 d (8.5) | 131.2 | 7.07 dd (8.5, 1.5) | 124.1 |
| 7′′′ | 7.65 d (15.5) | 146.9 | 7.66 d (16.0) | 147.2 |
| 8′′′ | 6.35 d (16.0) | 115.2 | 6.37 d (16.0) | 115.5 |
| 9′′′ | 168.7 | 168.4 | ||
| 3′′′-OCH3 | 3.91 s | 56.4 | ||
4. Discussion
This entry is adapted from the peer-reviewed paper 10.3390/plants10102032
References
- Xie, C.; Xie, Z.; Xu, X.; Yang, D. Persimmon (Diospyros kaki L.) leaves: A review on traditional uses, phytochemistry and pharmacological properties. J. Ethnopharmacol. 2015, 163, 229–240.
- Bei, W.; Zang, L.; Guo, J.; Peng, W.; Xu, A.; Good, D.A.; Hu, Y.; Wu, W.; Hu, D.; Zhu, X. Neuroprotective effects of a standardized flavonoid extract from Diospyros kaki leaves. J. Ethnopharmacol. 2009, 126, 134–142.
- Sakanaka, S.; Tachibana, Y.; Okada, Y. Preparation and antioxidant properties of extracts of Japanese persimmon leaf tea (kakinoha-cha). Food Chem. 2005, 89, 569–575.
- Sa, Y.S.; Kim, S.-J.; Choi, H.-S. The anticoagulant fraction from the leaves of Diospyros kaki L. has an antithrombotic activity. Arch. Pharmacal Res. 2005, 28, 667–674.
- Zhang, K.; Zhang, Y.; Zhang, M.; Gu, L.; Liu, Z.; Jia, J.; Chen, X. Effects of phospholipid complexes of total flavonoids from Persimmon (Diospyros kaki L.) leaves on experimental atherosclerosis rats. J. Ethnopharmacol. 2016, 191, 245–253.
- Kotani, M.; Matsumoto, M.; Fujita, A.; Higa, S.; Wang, W.; Suemura, M.; Kishimoto, T.; Tanaka, T. Persimmon leaf extract and astragalin inhibit development of dermatitis and IgE elevation in NC/Nga mice. J. Allergy Clin. Immunol. 2000, 106, 159–166.
- Thuong, P.T.; Lee, C.H.; Dao, T.T.; Nguyen, P.H.; Kim, W.G.; Lee, S.J.; Oh, W.K. Triterpenoids from the leaves of Diospyros kaki (persimmon) and their inhibitory effects on protein tyrosine phosphatase 1B. J. Nat. Prod. 2008, 71, 1775–1778.
- Matsuo, T.; Ito, S. The chemical structure of kaki-tannin from immature fruit of the persimmon (Diospyros kaki L.). Agric. Biol. Chem. 1978, 42, 1637–1643.
- Bawazeer, S.; Rauf, A. In vivo anti-inflammatory, analgesic, and sedative studies of the extract and naphthoquinone isolated from Diospyros kaki (persimmon). ACS Omega 2021, 6, 9852–9856.
- Yoshimura, M.; Mochizuki, A.; Amakura, Y. Identification of phenolic constituents and inhibitory activity of persimmon calyx and shiteito against tumor cell proliferation. Chem. Pharm. Bull. 2021, 69, 32–39.
- Wang, L.; Xu, M.L.; Rasmussen, S.K.; Wang, M.-H. Vomifoliol 9-O-α-arabinofuranosyl (1→ 6)-β-D-glucopyranoside from the leaves of Diospyros Kaki stimulates the glucose uptake in HepG2 and 3T3-L1 cells. Carbohydr. Res. 2011, 346, 1212–1216.
- Hitaka, Y.; Nakano, A.; Tsukigawa, K.; Manabe, H.; Nakamura, H.; Nakano, D.; Kinjo, J.; Nohara, T.; Maeda, H. Characterization of carotenoid fatty acid esters from the peels of the persimmon Diospyros kaki. Chem. Pharm. Bull. 2013, 61, 666–669.
- Simpson, D.S.; Oliver, P.L. ROS generation in microglia: Understanding oxidative stress and inflammation in neurodegenerative disease. Antioxidants 2020, 9, 743.
- Liu, Z.; Ren, Z.; Zhang, J.; Chuang, C.-C.; Kandaswamy, E.; Zhou, T.; Zuo, L. Role of ROS and nutritional antioxidants in human diseases. Front. Physiol. 2018, 9, 477.
- Kwon, J.; Hwang, H.; Selvaraj, B.; Lee, J.H.; Park, W.; Ryu, S.M.; Lee, D.; Park, J.-S.; Kim, H.S.; Lee, J.W. Phenolic constituents isolated from Senna tora sprouts and their neuroprotective effects against glutamate-induced oxidative stress in HT22 and R28 cells. Bioorganic Chem. 2021, 114, 105112.
- Romussi, G.; Bignardi, G.; Pizza, C.; De Tommasi, N. Constituents of cupuliferae, XIII: New and revised structures of acylated flavonoids from Quercus Suber L. Arch. Der Pharm. 1991, 324, 519–524.
- Li, H.-Z.; Song, H.-J.; Li, H.-M.; Pan, Y.-Y.; Li, R.-T. Characterization of phenolic compounds from Rhododendron alutaceum. Arch. Pharmacal Res. 2012, 35, 1887–1893.
- Jung, M.; Choi, J.; Chae, H.-S.; Cho, J.Y.; Kim, Y.-D.; Htwe, K.M.; Lee, W.-S.; Chin, Y.-W.; Kim, J.; Yoon, K.D. Flavonoids from Symplocos racemosa. Molecules 2015, 20, 358–365.
- Xu, J.; Wang, X.; Yue, J.; Sun, Y.; Zhang, X.; Zhao, Y. Polyphenols from acorn leaves (Quercus liaotungensis) protect pancreatic beta cells and their inhibitory activity against α-glucosidase and protein tyrosine phosphatase 1B. Molecules 2018, 23, 2167.
- Botirov, E.K. Flavonoids and phenolcarboxylic acids from Lamium album. Chem. Nat. Compd. 2019, 55, 1159–1160.
- Kawakami, K.; Shibukura, Y.; Kanno, T.; Furuki, T.; Aketa, S.; Hirayama, M. Identification of 2′′-galloylated flavonol 3-O-glycosides accumulating in developing leaves of Persimmon. Phytochem. Anal. 2011, 22, 403–410.
- Suktap, C.; Lee, H.K.; Amnuaypol, S.; Suttisri, R.; Sukrong, S. Wound healing effect of flavonoid glycosides from Afgekia mahidolae BL Burtt & Chermsir. Leaves. Rec. Nat. Prod. 2018, 12, 391–396.
- Isobe, T.; Ito, N.; Noda, Y. Minor flavonoids of Polygonum nodosum. Phytochemistry 1980, 19, 1877.
- Wan, C.; Yuan, T.; Cirello, A.L.; Seeram, N.P. Antioxidant and α-glucosidase inhibitory phenolics isolated from highbush blueberry flowers. Food Chem. 2012, 135, 1929–1937.
- Liao, C.-R.; Kuo, Y.-H.; Ho, Y.-L.; Wang, C.-Y.; Yang, C.-S.; Lin, C.-W.; Chang, Y.-S. Studies on cytotoxic constituents from the leaves of Elaeagnus oldhamii Maxim. in non-small cell lung cancer A549 cells. Molecules 2014, 19, 9515–9534.
- Wu, Z.-G.; Wei, W.; Xu, H.-Y.; Zheng, L.-L.; Ma, C.-M.; Wang, Y.-C. Constituents from the leaves of Tetraena mongolica and their protective activity in HEK 293t cells damaged by CdCl2. J. Nat. Prod. 2019, 82, 2707–2712.
- Kang, Y.-F.; Liu, C.-M.; Kao, C.-L.; Chen, C.-Y. Antioxidant and anticancer constituents from the leaves of Liriodendron tulipifera. Molecules 2014, 19, 4234–4245.
- Kwon, J.; Hiep, N.T.; Kim, D.-W.; Hong, S.; Guo, Y.; Hwang, B.Y.; Lee, H.J.; Mar, W.; Lee, D. Chemical constituents isolated from the root bark of Cudrania tricuspidata and their potential neuroprotective effects. J. Nat. Prod. 2016, 79, 1938–1951.
- Liu, Q.; Mu, Y.; An, Q.; Xun, J.; Ma, J.; Wu, W.; Xu, M.; Xu, J.; Han, L.; Huang, X. Total synthesis and anti-inflammatory evaluation of violacin A and its analogues. Bioorganic Chem. 2020, 94, 103420.
- Su, B.-N.; Kang, Y.-H.; Pinos, R.E.; Santarsiero, B.D.; Mesecar, A.D.; Soejarto, D.D.; Fong, H.H.; Pezzuto, J.M.; Kinghorn, A.D. Isolation and absolute stereochemistry of coussaric acid, a new bioactive triterpenoid from the stems of Coussarea brevicaulis. Phytochemistry 2003, 64, 293–302.
- Nakatani, M.; Miyazaki, Y.; Iwashita, T.; Naoki, H.; Hase, T. Triterpenes from Ilex rotunda fruits. Phytochemistry 1989, 28, 1479–1482.
- Lee, T.H.; Juang, S.H.; Hsu, F.L.; Wu, C.Y. Triterpene acids from the leaves of Planchonella duclitan (Blanco) Bakhuizan. J. Chin. Chem. Soc. 2005, 52, 1275–1280.
- Mimaki, Y.; Fukushima, M.; Yokosuka, A.; Sashida, Y.; Furuya, S.; Sakagami, H. Triterpene glycosides from the roots of Sanguisorba officinalis. Phytochemistry 2001, 57, 773–779.
- Fattorusso, E.; Lanzotti, V.; Taglialatela-Scafati, O.; Cicala, C. The flavonoids of leek, Allium porrum. Phytochemistry 2001, 57, 565–569.
- Yoshida, T.; Saito, T.; Kadoya, S. New acylated flavonol glucosides in Allium tuberosum Rottler. Chem. Pharm. Bull. 1987, 35, 97–107.
- Zehl, M.; Braunberger, C.; Conrad, J.; Crnogorac, M.; Krasteva, S.; Vogler, B.; Beifuss, U.; Krenn, L. Identification and quantification of flavonoids and ellagic acid derivatives in therapeutically important Drosera species by LC–DAD, LC–NMR, NMR, and LC–MS. Anal. Bioanal. Chem. 2011, 400, 2565–2576.
- Hossain, A.; Moon, H.K.; Kim, J.-K. Antioxidant properties of Korean major persimmon (Diospyros kaki) leaves. Food Sci. Biotechnol. 2018, 27, 177–184.
- Sun, L.; Zhang, J.; Lu, X.; Zhang, L.; Zhang, Y. Evaluation to the antioxidant activity of total flavonoids extract from persimmon (Diospyros kaki L.) leaves. Food Chem. Toxicol. 2011, 49, 2689–2696.
- Huang, S.-W.; Qiao, J.-W.; Sun, X.; Gao, P.-Y.; Li, L.-Z.; Liu, Q.-B.; Sun, B.; Wu, D.-L.; Song, S.-J. Secoiridoids and lignans from the leaves of Diospyros kaki Thunb. with antioxidant and neuroprotective activities. J. Funct. Foods 2016, 24, 183–195.
- Loizzo, M.R.; Said, A.; Tundis, R.; Hawas, U.W.; Rashed, K.; Menichini, F.; Frega, N.G.; Menichini, F. Antioxidant and antiproliferative activity of Diospyros lotus L. extract and isolated compounds. Plant Foods Hum. Nutr. 2009, 64, 264.
- Masuda, T.; Iritani, K.; Yonemori, S.; Oyama, Y.; Takeda, Y. Isolation and antioxidant activity of galloyl flavonol glycosides from the seashore plant, Pemphis acidula. Biosci. Biotechnol. Biochem. 2001, 65, 1302–1309.
- Fan, J.-P.; He, C.-H. Simultaneous quantification of three major bioactive triterpene acids in the leaves of Diospyros kaki by high-performance liquid chromatography method. J. Pharm. Biomed. Anal. 2006, 41, 950–956.
- Peng, L.; Zhao, M.; Li, H. Method development and validation for simultaneous determination of six flavonoids in rat eyes after oral administration of Diospyros kaki leaves extract by UPLC-MS/MS. Chem. Pharm. Bull. 2020, 69, 218–221.
