Engineering microbial consortia is an effective way for the removal of heavy metals and organic pollutants. In the study, we discussed the molecular and ecological mechanisms of engineering microbial consortia with a particular focus on metabolic cross-feeding within species and the transfer of metabolites. Besides, the advantages and limitations of top-down and bottom-up approaches of engineering microbial consortia were discussed, together with their applications in bioremediation.
- engineering microbial consortia
- bioremediation
- cross-feeding
- top-down engineering
- bottom-up engineering
1. Introduction
Microbes are ubiquitous organisms, found in air, soil, water, as well as animals, and plants [1]. They play vital roles in driving global biogeochemical cycles and have an immense impact on the survival, health, and development of mankind. A number of microbes have been isolated in laboratories that possess the ability to degrade organic pollutants and reduce or transform heavy metals [2]. However, the transforming efficiency of pollutants by a single species always declines when applied to in-site complex polluted sites [3]. Complex pollutants impose stress conditions on a single species and hinder their metabolism. In contrast, microbial consortia tend to show resistance and multifunctionality as varied species work together to efficiently utilize all forms of substrates [4][5].
Engineering microbial consortia may be an effective way to optimize the interaction within microorganisms and their environment and to ensure long-term stability. In the microbial communities, microbes compete for limited nutrients and consume metabolic products secreted by other species to gain fitness advantage [6]. It has been successfully applied in bioremediation of polluted sites, but also failed in some cases [7]. The metabolic interactions have a huge impact on the application of microbial consortia. The substances secreted by specific species can support or suppress the growth of other species, alter interactions between them and even influence the function of the whole community. Synthetic microbial consortia are defined as one that is created artificially by co-culturing of select (two or many) species under a (at least initially) well-defined media [8]. Unlike natural microbial consortia, it is feasible to reconfigure metabolic pathways and program social interactions of synthetic microbial consortia to obtain the desired function.
In this entry, we focused on the molecular mechanisms of microbial consortia, particularly metabolic cross-feeding between species ( Figure 1 ). We reviewed two main ways of engineering microbial consortia: top-down engineering and bottom-up engineering. Besides, we addressed important principles for engineering microbial consortia for the bioremediation of pollutants.
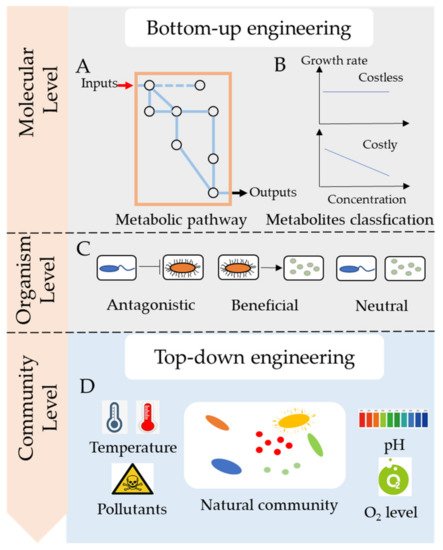
2. Microbial Cross-Feeding in Microbial Consortia
Microbial cross-feedings are common in natural environments [9] and microbes frequently exchange substances [10]. Many studies have reported that microbes significantly benefit from this exchange of metabolites [11]. In order to improve microbial consortia’s performance in bioremediation, it is essential to delineate the mechanisms of engineered microbial consortia on the molecular level by identifying metabolites and mechanisms of transfer.
Microbes commonly secrete metabolites across the membrane into the environment for utilization by other species. Several metabolites such as carbon and nitrogen resources, hydrogen (H 2), amino acids, vitamin, or growth factors may be exchanged between microbes [7]. These metabolites could be classified by molecule type, connection to metabolism, and fitness cost. Recently, more researches tended to focus on costly and costless metabolites, as they have different influences on the stability of microbial consortia [6][12]. For example, Bidirectional of costly metabolites may promote the growth of each species [13]. In contrast, unidirectional exchange of costly metabolites harms the secretors, and may lead the microbial consortia to collapse [14]. Besides, the exchange of costless metabolites doesn’t harm the secretors, instead benefits the receivers, thus, it causes no effect on the stability of microbial consortia. So, in this article, secreted metabolites are mainly classified into two categories, costly metabolites, and costless metabolites, depending on whether the secretion of metabolites causes a loss to the fitness of the organism.
Costly metabolites usually benefit the receivers but harm the producers as the increased secretion leads to reduced fitness. Creating a bidirectional exchange of costly metabolites could benefit the microbial consortia, which may help construct a stable community to improve bioremediation. In a study, an Escherichia coli mutant unable to synthesize methionine was co-cultured with Salmonella enterica ser. Typhimurium, which was able to receive metabolites from Escherichia coli [13]. After several generations, the Salmonella strain underwent mutations to excrete methionine, thereby aiding E. coli growth. That is to say, Salmonella gained fitness by receiving nutrients from enhanced E. coli growth, which helped Salmonella to overcome the fitness cost of high methionine excretion. Therefore, the bidirectional exchange of costly metabolites contributes to a reciprocal nutrient exchange that is beneficial for the whole community. However, there was little research on determining whether the metabolites of pollutants were costly or not. It causes risks of disrupting the functional microbial consortia when unknown species are added.
Microbes commonly secrete waste products with no fitness cost to the producer, that is, secretion does not alter the growth rate of the producer. It was proposed that costless metabolites can be a prominent driver of microbial interactions and influence the functions of microbial consortia [6]. The exchange of costless metabolites could offset competition for nutrients and yield stable specific partnerships, such as pollutants-degraders, which might help improve the bioremediation of pollutants. However, the cost of metabolite secretion and metabolic interactions of an organism may change in different environments [15] and are difficult to determine by experimental procedures. Using genome-scale models of metabolism, Pacheco et al. identified a large spectrum of costless metabolites [6].
3. How to Engineer Microbial Consortia towards Bioremediation
The engineering of microbial consortia is a complex task involving the assembly of different genera and species of microbes. There are two main approaches and several important principles of engineering microbial consortia towards bioremediation.
Bottom-up engineering is based on the metabolism and interactions between species. In the past, it was a challenge to predict and precisely manage microbial consortia; however, with the development of multi-omics and automation technology, many researchers have found ways to manipulate metabolic networks and microbial interactions [16][17][18].
Metabolism determines the nutrients that a species consumes and the metabolites that are excreted into the environment. There are several pathway models for the molecular mechanisms, such as glycolysis, the Kreb’s cycle, and glutaminolysis. Constraint-based methods, such as flux balance analysis, were also used for bottom-up engineering. Based on metabolic fluxes and interacting networks, it is possible to predict the desired output. Generally, the genome sequences of the strains in microbial consortia are needed to reconstruct the metabolic pathways and quantitative models are used to investigate the dynamics of the consortia [18]. Designing microbial consortia with defined social interactions is also an important part of bottom-up engineering. Microbial social interactions, such as competition and cooperation, are common in microbial communities and are essential in specifying ecosystem dynamics [19]. A model based on social-interaction programming can successfully predict the behaviors and dynamics of a community comprising of up to four species [20].
The core function of synthetic microbial consortia in the bioremediation of pollutants is the degradation of hazardous substances. Therefore, degraders are indispensable in a synthetic microbial consortium. However, there are still other important partner species with varied functions in the consortia that could contribute to the improvement of bioremediation ( Figure 2 ). When dealing with low water-soluble organic pollutants, the surfactant-producing function of microbial consortia needed to be considered. When the nutrient of polluted sites was poor, adding nitrogen fixers and carbon producers was important to provided degraders with enough nutrients for bioremediation. For instance, a Bacillus strain, part of a pyrene-degrading consortium, is unable to degrade pyrene, but can enhance the bioavailability of pyrene by producing biosurfactant [21]. A polycyclic aromatic hydrocarbons (PAHs)-degrading microbial consortium consisting of Pseudomonas and Actinobacteria strains also show emulsifying activities in the presence of PAHs, which notably helps the solubilization of PAHs during biodegradation [22].
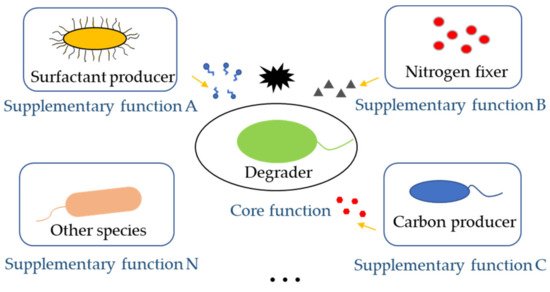
4. Engineering Microbial Consortia Promotes Bioremediation
| Pollutants | Microorganism | Bioremediation Efficiency | References |
|---|---|---|---|
| Pyrene | (Mycobacterium spp. PO1 and PO2, Novosphin-gobium pentaromativorans PY1, Ochrobactrum sp. PW1, and Bacillus sp. FW1 | Three-fold higher degradation rate for pyrene than the individual degrader. | [21] |
| Atrazine | Arthrobacter sp. DNS10, Bacillus subtilis DNS4 and Variovorax sp. DNS12, Arthrobacter sp. DNS9 | Removed 100% of atrazine at initial concentration of 100 mg/L, faster than single species. | [23] |
| PAHs | Rhodococcus sp., Acinetobacter sp., and Pseudomonas sp. | 100% degradation of Fl and Phe in sediment-free liquid medium after 4 weeks of growth. | [24] |
| Cr(VI) | Streptomyces sp. A5, A11, M7, MC1 | Removed 86% of Cr(VI) at initial concentration of 50 mg/kg in soil. | [25] |
| Lindane | Removed 46% of lindane at initial concentration of 25 mg/kg in soil. | ||
| Cd | Bacillus sp. strain H9, Ralstonia eutropha JMP134 | Removed 42% of phenanthrene at initial concentration of 24 mg/L. | [26] |
| 2,4-D | Removed 100% of 2,4-D at initial concentration of 500 mg/L. |
4.1. Organic Pollutants
4.2. Heavy Metals
4.3. Complex Pollution
5. Conclusions
This entry is adapted from the peer-reviewed paper 10.3390/w13202928
References
- Whitman, W.B.; Coleman, D.C.; Wiebe, W.J. Prokaryotes: The Unseen Majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583.
- Kanaly, R.A.; Harayama, S. Biodegradation of High-Molecular-Weight Polycyclic Aromatic Hydrocarbons by Bacteria. J. Bacteriol. 2000, 182, 2059–2067.
- Sandrin, T.R.; Maier, R.M. Impact of Metals on the Biodegradation of Organic Pollutants. Environ. Health Perspect. 2003, 111, 1093–1101.
- Brisson, V.L.; West, K.A.; Lee, P.K.H.; Tringe, S.G.; Brodie, E.L.; Alvarez-Cohen, L. Metagenomic Analysis of a Stable Trichloroethene-Degrading Microbial Community. ISME J. 2012, 6, 1702–1714.
- Hansen, S.K.; Rainey, P.B.; Haagensen, J.A.J.; Molin, S. Evolution of Species Interactions in a Biofilm Community. Nature 2007, 445, 533–536.
- Pacheco, A.R.; Moel, M.; Segrè, D. Costless Metabolic Secretions as Drivers of Interspecies Interactions in Microbial Ecosystems. Nat. Commun. 2019, 10, 1–12.
- Seth, E.C.; Taga, M.E. Nutrient Cross-Feeding in the Microbial World. Front. Microbiol. 2014, 5, 1–6.
- Großkopf, T.; Soyer, O.S. Synthetic Microbial Communities. Curr. Opin. Microbiol. 2014, 18, 72–77.
- Ponomarova, O.; Patil, K.R. Metabolic Interactions in Microbial Communities: Untangling the Gordian Knot. Curr. Opin. Microbiol. 2015, 27, 37–44.
- Giri, S.; Shitut, S.; Kost, C. Harnessing Ecological and Evolutionary Principles to Guide the Design of Microbial Production Consortia. Curr. Opin. Biotechnol. 2020, 62, 228–238.
- Hynes, W.F.; Chacón, J.; Segrè, D.; Marx, C.J.; Cady, N.C.; Harcombe, W.R. Bioprinting Microbial Communities to Examine Interspecies Interactions in Time and Space. Biomed. Phys. Eng. Express 2018, 4, 25–33.
- Harcombe, W.R.; Chacón, J.M.; Adamowicz, E.M.; Chubiz, L.M.; Marx, C.J. Evolution of Bidirectional Costly Mutualism from Byproduct Consumption. Proc. Natl. Acad. Sci. USA 2018, 115, 12000–12004.
- Harcombe, W. Novel Cooperation Experimentally Evolved between Species. Evolution 2010, 64, 2166–2172.
- Van Tatenhove-Pel, R.J.; de Groot, D.H.; Bisseswar, A.S.; Teusink, B.; Bachmann, H. Population Dynamics of Microbial Cross-Feeding Are Determined by Co-Localization Probabilities and Cooperation-Independent Cheater Growth. ISME J. 2021, 15, 3050–3061.
- Tarnita, C.E. The Ecology and Evolution of Social Behavior in Microbes. J. Exp. Biol. 2017, 220, 18–24.
- Löffler, F.E.; Edwards, E.A. Harnessing Microbial Activities for Environmental Cleanup. Curr. Opin. Biotechnol. 2006, 17, 274–284.
- Gude, S.; Taga, M.E. Multi-Faceted Approaches to Discovering and Predicting Microbial Nutritional Interactions. Curr. Opin. Biotechnol. 2020, 62, 58–64.
- Embree, M.; Liu, J.K.; Al-Bassam, M.M.; Zengler, K. Networks of Energetic and Metabolic Interactions Define Dynamics in Microbial Communities. Proc. Natl. Acad. Sci. USA 2015, 112, 15450–15455.
- Faust, K.; Raes, J. Microbial Interactions: From Networks to Models. Nat. Rev. Microbiol. 2012, 10, 538–550.
- Kong, W.; Meldgin, D.R.; Collins, J.J.; Lu, T. Designing Microbial Consortia with Defined Social Interactions. Nat. Chem. Biol. 2018, 14, 821–829.
- Wanapaisan, P.; Laothamteep, N.; Vejarano, F.; Chakraborty, J.; Shintani, M.; Muangchinda, C.; Morita, T.; Suzuki-Minakuchi, C.; Inoue, K.; Nojiri, H.; et al. Synergistic Degradation of Pyrene by Five Culturable Bacteria in a Mangrove Sediment-Derived Bacterial Consortium. J. Hazard. Mater. 2018, 342, 561–570.
- Isaac, P.; Martínez, F.L.; Bourguignon, N.; Sánchez, L.A.; Ferrero, M.A. Improved PAHs Removal Performance by a Defined Bacterial Consortium of Indigenous Pseudomonas and Actinobacteria FromPatagonia, Argentina. Int. Biodeterior. Biodegrad. 2015, 101, 23–31.
- Zhang, Y.; Cao, B.; Jiang, Z.; Dong, X.; Hu, M.; Wang, Z. Metabolic Ability and Individual Characteristics of an Atrazine-Degrading Consortium DNC5. J. Hazard. Mater. 2012, 237–238, 376–381.
- Yu, S.H.; Ke, L.; Wong, Y.S.; Tam, N.F.Y. Degradation of Polycyclic Aromatic Hydrocarbons by a Bacterial Consortium Enriched from Mangrove Sediments. Environ. Int. 2005, 31, 149–154.
- Polti, M.A.; Aparicio, J.D.; Benimeli, C.S.; Amoroso, M.J. Simultaneous Bioremediation of Cr(VI) and Lindane in Soil by Actinobacteria. Int. Biodeterior. Biodegrad. 2014, 88, 48–55.
- Roane, T.M.; Josephson, K.L.; Pepper, I.L. Dual-Bioaugmentation Strategy to Enhance Remediation of Cocontaminated Soil. Appl. Environ. Microbiol. 2001, 67, 3208–3215.
- HuiJie, L.; CaiYun, Y.; Yun, T.; GuangHui, L.; TianLing, Z. Using Population Dynamics Analysis by DGGE to Design the Bacterial Consortium Isolated from Mangrove Sediments for Biodegradation of PAHs. Int. Biodeterior. Biodegrad. 2011, 65, 269–275.
- Brune, K.D.; Bayer, T.S. Engineering Microbial Consortia to Enhance Biomining and Bioremediation. Front. Microbiol. 2012, 3, 1–6.
- Mosharaf, M.K.; Tanvir, M.Z.H.; Haque, M.M.; Haque, M.A.; Khan, M.A.A.; Molla, A.H.; Alam, M.Z.; Islam, M.S.; Talukder, M.R. Metal-Adapted Bacteria Isolated from Wastewaters Produce Biofilms by Expressing Proteinaceous Curli Fimbriae and Cellulose Nanofibers. Front. Microbiol. 2018, 9, 1–17.
- Smith, W.L. Hexavalent Chromium Reduction and Precipitation by Sulphate-Reducing Bacterial Biofilms. Environ. Geochem. Health 2001, 23, 297–300.
- Dhal, B.; Thatoi, H.N.; Das, N.N.; Pandey, B.D. Chemical and Microbial Remediation of Hexavalent Chromium from Contaminated Soil and Mining/Metallurgical Solid Waste: A Review. J. Hazard. Mater. 2013, 250–251, 272–291.
- Shen, H.; Wang, Y.T. Simultaneous Chromium Reduction and Phenol Degradation in a Coculture of Escherichia Coli ATCC 33456 and Pseudomonas Putida DMP-1. Appl. Environ. Microbiol. 1995, 61, 2754–2758.
