Metabolomics is one of the most complex omics tools; the added complexity stems from the fact that there is not one set of metabolites making up the metabolome. Indeed, the metabolome is highly dynamic and diverse with metabolites originating external to the host and metabolites that are synthesised endogenously. This added complexity means that the metabolome can be informative both regarding external exposures including dietary intake and on the metabolic phenotype.
- metabolomics
- dietary intake
- metabolites
1. Introduction
Traditionally, nutritional epidemiology is the study of the relationship between diet and health and disease in humans at the population level. In this case, food intake is the exposure of interest, and it has been studied in relation to a wide range of health and disease outcomes. Classically, epidemiologists examine how dietary intake impacts the occurrence of disease through collection of data in a large population and comparing groups within this population for disease incidence. Statistical approaches are used to estimate the extent to which the exposure influences the risk of disease in the population. The measures are usually associations, and causal or mechanistic underpinnings are difficult if not impossible to establish in such approaches. Notwithstanding this, nutritional epidemiology studies play an important role in the development of policies on diet, health and disease and in the guidance of fortification policies [1]. With the widespread uptake of omic technologies, nutritional epidemiology has moved from a “black box” approach to a systems approach where genomics, metabolomics and proteomics are providing novel insights into the interplay between diet and health [2,3]. Due to the close interplay between food and metabolism, the application of metabolomics in nutritional epidemiology has been particularly successful.
2. Addressing Limitations of Self-Reported Dietary Intake Data
Nutritional epidemiological studies with large numbers of participants have often relied on self-reported instruments to assess dietary intake. Although these instruments such as food frequency questionnaires (FFQs) have been useful to estimate habitual dietary intakes and characterize dietary patterns in free-living populations, it is well documented that these instruments are prone to random and systematic errors. Examples of such errors include underestimated energy intake, recall inaccuracies or biases and difficulty in assessment of portion sizes [4,5,6]. These errors can result in reduced power and underestimated or even inflated associations which in turn partly contributes to the inconsistencies in results in nutritional epidemiology [7,8]. Other instruments such as 24-hr recalls are also subject to recall inaccuracies and random within-person variabilities. The diet record method, which is considered the most accurate self-reported dietary assessment method, is burdensome and expensive and thus often infeasible in large epidemiological studies. To address some of the concerns surrounding self-reported dietary data, the potential role of dietary biomarkers has emerged. While classical biomarkers for salt, protein and energy intake have existed for years, the emergence of metabolomics has resulted in the expansion of dietary biomarkers to include biomarkers for specific foods and dietary patterns [9,10,11]. While the potential of classical biomarkers for correcting self-reported data is well-established, there is emerging evidence that novel food and nutrient biomarkers that are discovered agnostically through metabolomic profiling can also be employed to correct self-reported data.
Lampe and colleagues illustrated the potential of candidate biomarkers for dietary assessment in terms of nutrient intake [12]. Using a series of dietary biomarkers including carotenoids, tocopherols, folate, and vitamin B12, the authors demonstrated that these biomarkers performed as well as the established biomarkers of energy (doubly labelled water) and protein (urinary nitrogen) in estimating nutrient intake. These and other biomarkers have the potential to be used to calibrate self-reported data in large studies [12]. Extension to include the calibrated intakes into regression models allows diet–disease associations to be explored [13].
A study of 2208 men and women identified 65 metabolites that were associated with at least one of the three dietary patterns examined (AHEI, DASH and MDS) [23]. A shared signature of 24 metabolites was associated with all three healthy dietary patterns. Many more examples exist where dietary patterns are correlated to metabolites, and these lay the foundations for the potential of biomarkers to correct self-reported data in order to obtain dietary patterns.
Using panels of biomarkers to assign individuals into dietary patterns with no reliance on self-reported data is also attractive. While further work is needed to develop this concept, there are some examples that demonstrate the potential and support additional work in this area. Employing a controlled intervention design, Garcia-Perez and colleagues developed a model based on urinary metabolomics data that could classify individuals into dietary patterns, which was validated in two separate population groups [25]. Using urinary metabolomic data only, individuals were classified into four dietary patterns: replication was achieved in a separate group with good reproducibility over four time points [26]. Further work is needed to expand these approaches and develop them for use in a range of population groups.
3. Using Metabolites to Inform about Metabolic Processes
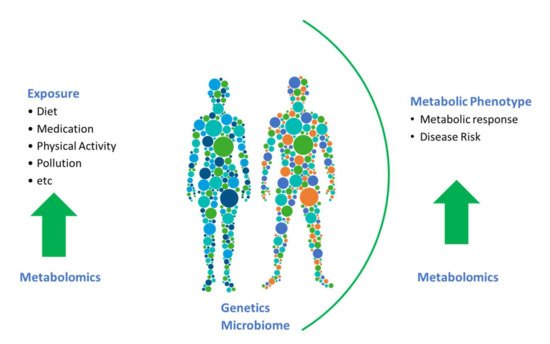
To date, the majority of studies in nutritional epidemiology have focused on associations between biomarkers and dietary-intake data, the ultimate challenge is now to move beyond this. Biomarkers, and in particular, endogenous biomarkers, have much more to offer (see Figure 1 ). To a certain extent, the full potential of metabolomics has been hindered by a focus on associating metabolomics data with dietary data for identification of food-intake biomarkers. We need to embrace linking metabolites to their physiological roles and endogenous metabolic pathways and networks. There is a need to assess the impact of diet on such metabolic pathways and physiological processes, and determine how diet impacts changes in levels of metabolites and consequently health and disease outcomes in prospective cohorts. In the era of precision nutrition, it is also critical to identify and examine biomarkers that reflect the effects of gene–diet or microbiota–diet interactions to help identify subpopulations that may benefit from targeted dietary advice.
A recent study profiled over 1000 metabolites in approximately 11,000 individuals to examine metabolic pathways associated with and across 27 incident noncommunicable diseases (NCDs) [27]. A total of 420 metabolites were shared between at least two NCDs, and metabolic pathway analysis revealed a number of key pathways common across the diseases. A web server was developed, incorporating the results to enable future analysis at the metabolite and pathway level.
With the emergence of rich datasets incorporating multiomics data, the integration of metabolomic data with other omic data is an important approach to obtain functional information. Integration of the blood metabolome with deep immunophenotyping in 500 individuals identified metabolite features associated with eight categories of host factors, including baseline immune parameters and immune cytokine response [28]. Using genome-wide association analysis, a comprehensive landscape of genetic regulation on metabolism was identified. A number of metabolite quantitative trait loci (mQTLs) were identified, and further Mendelian Randomisation analysis revealed that one locus associated with arachidonic acid was causally associated with Crohn’s disease. This study is a pertinent example of how integration of omic datasets provides new insights into the interplay between genes, metabolites and disease risk.
In a UK study of 10,806 participants a total of 66 metabolites were significantly associated with the MDS [22]. These metabolites were combined into a metabolite score which was estimated to explain 37.2% of the inverse association of the Mediterranean diet score with Homeostatic Model of Insulin Resistance (HOMA-IR). The authors suggest that the metabolites including acylcarnitines, sphingolipids and phospholipids act as mediators, and propose their involvement in pathways linking diet to disease risk. This study highlights the potential use of metabolites and is an elegant example of the information we can harness from them.
This entry is adapted from the peer-reviewed paper 10.3390/metabo11100709
