Rbm24 gene exhibits strongly conserved expression in the somites, heart, lens and otic vesicle of all vertebrate embryos. It is involved in different aspects of post-transcriptional regulation of gene expression, including pre-mRNA alternative splicing, mRNA stability and polyadenylation. Knockout of this gene in mice and zebrafish causes severe developmental defects, such as heart malformations, cataract formation and hearing deficits.
- Post-transcriptional regulation
- alternative splicing
- cytoplasmic polyadenylation
- heart
- cataract
- cell differentiation
- muscle
- inner ear
- RBP
- Rbm24
1. Definition
Rbm24 (RNA-binding motif protein 24) is an evolutionarily conserved RBP that contains a single RNA recognition motif (RRM) at its N-terminal region [13–15]. It exhibits strongly restricted tissue-specific expression patterns during vertebrate development [16], and emerges as an important regulator of cellular differentiation and a potential factor implicated in human disease.
2. Introduction
RNA-binding proteins (RBPs) play key roles in the post-transcriptional regulation of gene expression in a variety of biological processes. Vertebrate cells express hundreds to thousands of RBPs that display unique binding activity to their RNA targets and specific interaction with other protein partners. They control RNA metabolism at multiple levels, from alternative splicing, to transport, subcellular localization, stability, polyadenylation, and translation [1]. Thus, protein–RNA interactions are critical for maintaining the homeostasis of protein synthesis during early development and in adult life. Indeed, large-scale analyses of protein expression regulation have made the unprecedented finding that protein abundances within the proteome and mRNA levels within the transcriptome are entirely uncoupled in many conditions [2,3]. This highlights the importance of additional regulations by the ribonome, which embodies mRNAs and associated RBPs within a rich ribonucleoprotein (RNP) infrastructure in the cell [4]. Due to the crucial roles exerted by the processes downstream of transcription in the spatiotemporal control of gene expression, it is not surprising that a growing number of human diseases, such as neurodegenerative disorders and various types of cancers, are associated with RNAs and RBPs [5–9]. Therefore, RBPs may have the potential to be used as therapeutic targets in many diseased conditions [10–12].
Although no mutation of the human RBM24 gene has been associated with any disease up today, deficiency in its expression level could be the cause of congenital disorders, such as cardiomyopathy, myopathy, or blindness, which affect the normal function of related tissues where crucial roles of this gene have been demonstrated in different animal models. It seems that vertebrate Rbm24 could be involved in nearly all aspects of post-transcriptional regulation. Most importantly, it functions as a key factor that regulates alternative splicing to establish the contractile function in developing cardiac and skeletal muscles [17–19], and plays an important role in cytoplasmic polyadenylation (CPA) to ensure the accumulation of crystallin proteins during lens fiber cell terminal differentiation [20]. There is thus accumulating evidence that Rbm24 acts as a multifaceted regulator to initiate cell differentiation through distinct mechanisms, which may vary in a tissue-specific and even a stage-specific manner. Moreover, Rbm24 presents almost the same characteristics in RNA and protein interactions, and often performs similar functions as the closely related ancient paralog Rbm38 (RNPC1) in regulating cell cycle progression and apoptosis [21,22], adding more complexity to its functional implication in various cellular processes. Mechanistic insights on how Rbm24 controls gene expression are beginning to be elucidated using in vivo and in vitro approaches. However, a number of important questions remain unanswered and await further investigation, such as its dynamic subcellular localization and tissue-specific function, the modulation of its activity through interaction with other partners, and its potential implication in human pathologies.
3. Rbm24 Functional Domains
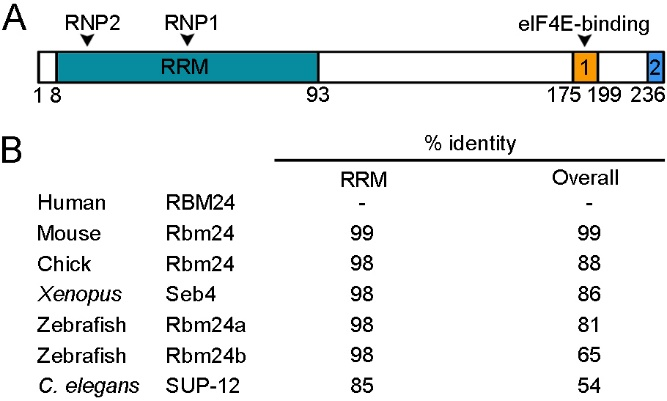
Rbm24 proteins display deep evolutionary conservation. The RRM is located at the N-terminal half and contains two characteristic consensus RNP sequences, RNP1 and RNP2 [23]. This domain shows extremely high levels of sequence identity among different species, ranging from a nematode to human (Figure 1).
Figure 1. Rbm24 is a highly conserved RNA-binding protein (RBP). (A) Schematic representation of human RBM24 protein domains. The amino-terminal half contains a canonical RRM with two consensus RNP sequences (RNP1 and RNP2), and the C-terminal region contains two conserved domains (1 and 2), including an eIF4E-binding motif. Amino acid positions are indicated below. (B) Identity in the overall sequence and in the RRM between human RBM24 and Rbm24 proteins from other species. Human RBM24, NM_001143942.2; mouse Rbm24, NM_001081425.1; chick Rbm24, NM_001012863.3; Xenopus laevis Seb4, NM_001087526.1; zebrafish Rbm24a, NM_212865.1; zebrafish Rbm24b, NM_001328141.1; C. elegans SUP-12, NM_076273.5.
In addition, it is almost identical to the RRM of Rbm38, such that both Rbm24 and Rbm38 prefers similar GU-rich sequences in target mRNAs [21,24]. Although the sequence outside the RRM is relatively divergent, at least two conserved domains can be identified in the C-terminal half of vertebrate Rbm24 and Rbm38 [25]. In particular, a motif close to the extreme C-terminus, which contains a serine residue (serine 181 in Rbm24 and serine 195 in Rbm38), interacts with eukaryote initiation factor 4E (eIF4E) and prevents it from binding to the 5’-cap of mRNAs [26,27]. However, at least in several cancer cell lines, phosphorylation of the serine residue by glycogen synthase kinase 3 (GSK3) prevents the interaction with eIF4E and converts Rbm24 or Rbm38 into an activator of mRNA translation [28]. A second conserved motif is the extreme C-terminus, whose implication in the Rbm24 function is not clear. The C-terminal half of Rbm24 also interacts with a number of other partners, but the exact motif implicated has not been determined yet, and the interaction is often assisted by the RRM. For example, both the N-terminal and C-terminal regions of Rbm24 are required for interaction with Stk38 kinase [29]. Although the N-terminal region of zebrafish Rbm24a does not directly interact with cytoplasmic polyadenylation element-binding protein 1b (Cpeb1b) and cytoplasmic poly(A)-binding protein 1l (Pabpc1l), its absence decreases the capacity of the C-terminal region to interact with these partners [20]. Thus, Rbm24 displays biochemical and functional conservation with respect to Rbm38, and its functionality in different cellular processes may be regulated through interaction with specific protein partners.
4. Rbm24 Displays Restricted Expression Patterns in Developing Vertebrate Embryos
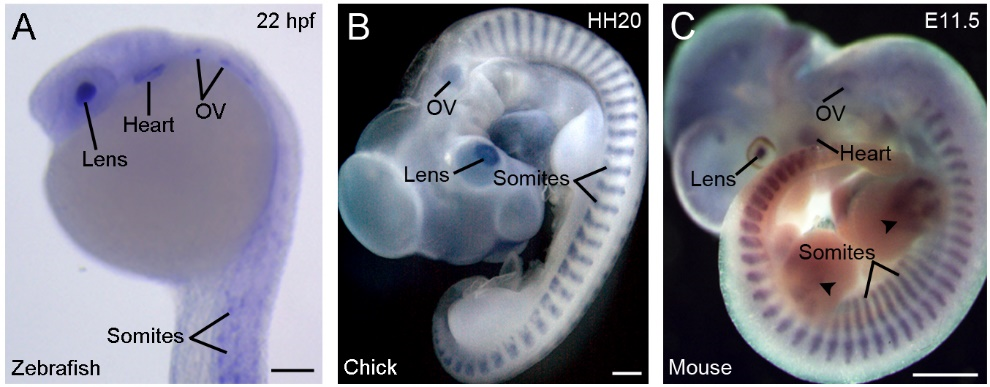
Another strikingly conserved aspect of vertebrate Rbm24 is the highly restricted developmental expression patterns (Figure 2).
Figure 2. Conserved and restricted expression patterns of Rbm24 gene in vertebrate embryos. (A) The expression of rbm24a in a zebrafish embryo at 22 hpf (hours post-fertilization). (B) A chick embryo at stage HH20 (3 days). (C) A mouse embryo at E11.5 [16]. Arrowheads indicate Rbm24 expression in limb muscles. In all vertebrates, Rbm24 is expressed in the somites, heart, lens, and otic vesicle (OV). Scale bars: (A) 100 µm; (B) 1 mm; and (C) 1 mm.
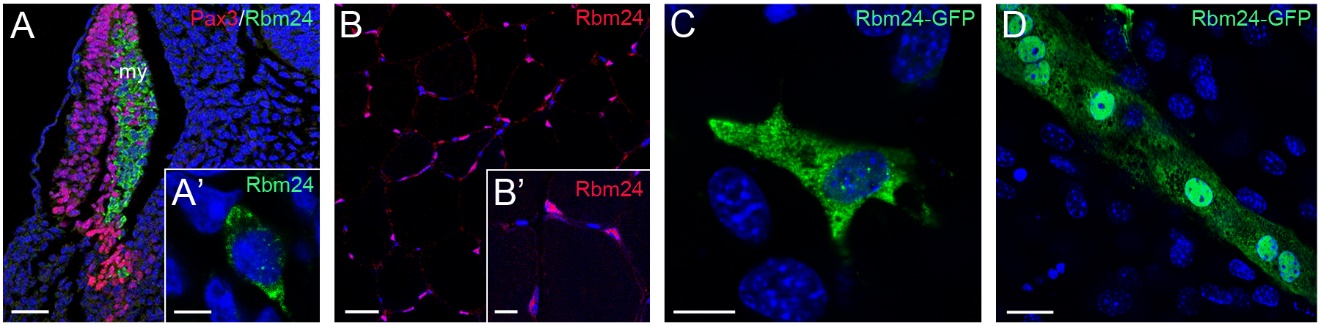
During gastrulation, Xenopus rbm24, also called XSeb4 or MTG1, is mainly expressed in the dorso-lateral mesoderm regions flanking the axial midline, which mostly overlap with the expression domains of MyoD [13,30]. During organogenesis, analyses by in situ hybridization and immunofluorescence staining indicate that Rbm24 is expressed in the somites, heart, and head sensory organs in different vertebrate embryos, including zebrafish, Xenopus, chick, and mouse [13,14,16,31–36]. Zebrafish genome contains two rbm24 paralogs, rbm24a and rbm24b. The sites of rbm24a expression are identical as those of other vertebrate Rbm24, whereas rbm24b is mainly expressed in the somites and heart, but not in head sensory organs [33,34]. Within each tissue, Rbm24 is generally expressed in a subset of cells undergoing differentiation. The cellular localization of the Rbm24 protein in different tissues has been analyzed in more detail during mouse embryonic development [16,36]. Interestingly, Rbm24 displays dynamic subcellular localization during muscle cell development. In the myotome of mouse embryos and in murine C2C12 cells it is first accumulated in the cytoplasm of MyoD-positive myoblasts entering into the differentiation program, but not in Pax3-positive muscle progenitor cells, while in mature myotubes it is mainly present in the nucleus (Figure 3).
Figure 3. Dynamic subcellular localization of Rbm24 protein during muscle cell differentiation. (A) Immunofluorescence staining of a mouse embryonic section at E11.5 shows Rbm24 expression in differentiating myoblasts within the myotome (my), but not in Pax3-positive premyogenic progenitors. Some Pax3-expressing cells are intermingled with Rbm24-positive cells, but they do not yet express Rbm24 [16]. (A’) Higher magnification shows the punctate cytoplasmic staining of Rbm24 in a myoblast. (B) Immunofluorescence staining of Rbm24 in adult mouse muscles. (B’) Higher magnification shows nuclear localization of Rbm24. (C) Localization of Rbm24-GFP in the cytoplasm of murine C2C12 myoblasts. (D) Strong nuclear and weak cytoplasmic localization of Rbm24-GFP in differentiated C2C12 myotubes. Nuclei are stained with DAPI. Scale bars: (A) 50 µm; (A’) 10 µm; (B) 20 µm; (B’) 10 µm; (C) 10 µm; and (D) 10 µm.
A cytoplasmic localization of the Rbm24 protein is also detected in different head sensory organs [36]. In the developing lens, it is restricted to differentiating fiber cells in the posterior and equatorial regions; in the otic vesicle, it co-localizes with Myo7A in inner ear hair cells; in the olfactory epithelium, it is expressed in fate-committed neuronal precursors and terminally differentiated olfactory receptor neurons. Thus, the predominant localization of Rbm24 in the cytoplasmic compartment of different cell types closely correlates with their entry into the process of differentiation. Detailed Rbm24 expression and subcellular localization will be further described in sections addressing its tissue-specific function. Herein we discuss the regulatory roles of Rbm24 and the consequences of its dysfunction in development and disease (Table 1).
Table 1. Potential Rbm24 functions in development and disease.
|
Tissue or Disease |
Post-Transcriptional Regulation |
Role in Development or Disease |
|
Skeletal muscle |
Muscle-specific pre-mRNA splicing, mRNA stability |
Myogenic differentiation, somitogenesis, and sarcomere organization [16,17,30,34,37] |
|
Cardiac muscle |
Muscle-specific pre-mRNA splicing, mRNA stability and translation |
Heart development, sarcomere assembly, and cardiac contractility [14,17,19,33,38] |
|
Lens |
Cytoplasmic polyadenylation, mRNA stability |
Lens fiber cell differentiation, and lens transparency [20,39] |
|
Inner ear/Neuromasts |
mRNA stability |
Hair cell morphogenesis and differentiation [40] |
|
Olfactory epithelium |
Unknown (cytoplasmic localization in neuronal cells) |
Unknown |
|
Blastula/Gastrula |
Unknown |
Germ layer formation [30] |
|
Prostate cancer |
Interaction with miR-106a-5p |
Inhibition of tumorigenesis [41] |
|
Nasopharyngeal carcinoma |
Interaction with miR-25 |
Inhibition of cell proliferation, migration and invasion [42] |
|
Hirschsprung disease |
Interaction with MIR143HG |
Proliferation or migration of enteric neural crest cells, and gangliogenesis [43] |
|
Familial dysautonomia |
Increased recognition of the mutated 5’ splicing site in IKBKAP gene by U1 snRNP |
Possible protective role against the aberrant splicing of the mutated gene [44] |
|
Hepatitis A and B |
Pregenomic RNA packaging and replication |
Possible host factor for the viruses [45–47] |
This entry is adapted from the peer-reviewed paper 10.3390/cells9081891