Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Others
|
Biotechnology & Applied Microbiology
Nostocyclopeptides (Ncps) are a small class of bioactive nonribosomal peptides thus far identified only in cyanobacteria of the genus Nostoc. They are composed of six-seven amino acid residues and contain a unique imino linkage formed between C-terminal aldehyde and an N-terminal amine group of the conserved tyrosine. Nostocyclopeptides occur both in cyclic and linear form.
- 20S proteasome inhibitors
- cyanobacteria
- Nostoc
- nostocyclopeptides
1. Introduction
The 26S proteasome is a large (2.4 MDa), multifunctional and ATP-dependent enzymatic complex with chymotrypsin-like (CT-L), trypsin-like (T-L), and caspase-like (C-L) activities [1][2][3][4]. In eukaryotic organisms, it recognizes and degrades proteins with covalently attached ubiquitin (8.5 kDa protein) [5][6]. The 26S proteasome is composed of a 20S barrel-shaped core particle (700 kDa) responsible for proteolytic activity and one or two 19S (890 kDa) regulatory subunits with ubiquitin-binding sites [3][4][7]. The 20S proteasome also occurs as a free complex that degrades proteins in the ubiquitin-independent pathway [8][9]. In humans, the dysfunction of this proteolytic machinery leads to changes in protein profile and, ultimately, to serious health problems. Therefore, proteasome regulators are explored as promising therapeutic agents for a range of diseases (e.g., cancer, autoimmune disorders, inflammation, malaria) [10][11][12]. The majority of the known 20S proteasome inhibitors belongs to peptide-based structures such as peptide aldehydes, boronates, epoxyketones, or peptide vinyl sulfones [13][14][15]. Some of the active compounds are of natural origin. Leupeptin, isolated from several strains of Gram-positive bacteria of the order Actinomycetales, inhibits T-L activity of the 20S proteasome [16]. Tyropeptin A, a peptide aldehyde produced by the soil Streptomycetales of the genus Kitasatospora, strain MK993-dF2, inhibits mainly CT-L activity [17][18]. Marine fungus Peicillium fellutanum is a producer of fellutamide B, a strong inhibitor of CT-L activity (IC50 9.4 nM) with mild effects on T-L (IC50 2.0 μM) and C-L (IC50 1.2 μM) activities [19]. The proteasome inhibition within the nanomolar to the micromolar range of IC50 was also documented for metabolites isolated from cyanobacteria Symploca sp., Scytonema hofmannii, and Nostoc.
In our preliminary studies, fractions from Nostoc edaphicum CCNP1411 containing
nostocyclopeptides (Ncps) inhibited the chymotrypsin-like activity of the 20S proteasome.
Ncps constitute a small group of nonribosomal peptides solely produced by cyanobacteria
of the genus Nostoc. The biological activity of the peptides was reported in several studies.
According to Golakoti et al. [20], Ncp-A1 and Ncp-A2 have cytotoxic activity against
human colorectal adenocarcinoma (LoVo) and human nasopharyngeal (KB) cell lines
(IC50 ca. 1 µM). Another nostocyclopeptide variant, Ncp-M1, was shown to inhibit the
transport of toxic microcystin-LR and nodularin into hepatocytes by blocking organic
anion transporter polypeptides, OATP1B1, and OATP1B3. These polypeptides are also
overexpressed in cancer cells [21][22]. The role of Ncp-M1 and its analogs as antitumor
agents and as tools to study membrane transport was proposed [21][23][24].
nostocyclopeptides (Ncps) inhibited the chymotrypsin-like activity of the 20S proteasome.
Ncps constitute a small group of nonribosomal peptides solely produced by cyanobacteria
of the genus Nostoc. The biological activity of the peptides was reported in several studies.
According to Golakoti et al. [20], Ncp-A1 and Ncp-A2 have cytotoxic activity against
human colorectal adenocarcinoma (LoVo) and human nasopharyngeal (KB) cell lines
(IC50 ca. 1 µM). Another nostocyclopeptide variant, Ncp-M1, was shown to inhibit the
transport of toxic microcystin-LR and nodularin into hepatocytes by blocking organic
anion transporter polypeptides, OATP1B1, and OATP1B3. These polypeptides are also
overexpressed in cancer cells [21][22]. The role of Ncp-M1 and its analogs as antitumor
agents and as tools to study membrane transport was proposed [21][23][24].
2. Results and Discussion
Thus far, the presence of Ncps was reported in five strains of Nostoc isolated from different habitats [20][21][22][23][24]. This includes two Baltic strains: XSPORK 13A producing the cyclic Ncp-M1 [21] and CCNP1411 producing 10 other Ncps variants [24]. The putative structures of the five linear and five cyclic Ncps variants produced by CCNP1411 were elucidated based on mass spectra fragmentation patterns [24]. Two of the cyclic forms, Ncp-A1 and Ncp-A2, enclosed by imino linkage between the N-terminal amine group of conserved Tyr and C-terminal aldehyde group of Leu or Phe, were previously identified in Nostoc sp. ATCC53789 isolated from lichen [20]. In position 6 of the Ncps from CCNP1411, 4-methylproline (MePro) or Pro is present, while Ile or Val is in position 4 (Figure 1). In the current study, we were able to isolate 6 out of 10 Ncps produced by CCNP1411 (Table 1): three cyclic variants (Ncp-A1, Ncp-A2, and Ncp-E2), two linear aldehyde forms of the cyclic variants (Ncp-A2-L and Ncp-E2-L), and the six-amino acid peptide Ncp-E4-L lacking the aldehyde group in the C-terminus. In the case of four other Ncps produced by CCNP1411 (Ncp-A1-L, Ncp-E1, Ncp-E1-L, Ncp-E3), their purity and/or quantities were not sufficient for inclusion in the study. Ncp-A2-L (Figure 1) was the only variant obtained in sufficient amounts for NMR analyses. The 1H NMR spectrum of Ncp-A2-L displayed a typical pattern of a peptide. The COSY, TOCSY, and HMBC experiments allowed for the identification of the residues in Ncp-A2-L as Tyr, Gly, Gln, Ile, Ser, MePro, and phenylalaninal (Phe-H) (Table 2, Figure 1, Figures S1–S5). The amino acid sequence was confirmed by TOCSY data. The presence of two aromatic amino acid residues was recognized by the signals occurring in the aromatic region of the spectrum (δH 6.78–7.26 ppm). One of them was identified as tyrosine-based on the AA’BB’ spin system between the aromatic protons (Tyr-H5/5′ and Tyr-H6/6′, JH, H = 8.0 Hz). The second aromatic residue was identified as phenylalanine based on the TOCSY interaction between 34, 35, and 36 protons and the HMBC correlation from two diastereotopic methylene protons 32a (δH 2.57 ppm) and 32b (δH 2.98 ppm) to the aromatic 34/34′ carbons (Figures S3 and S5). The 4-methyl group of the proline residue was identified based on the 1H NMR doublet signal at δ 0.82 ppm (protons 29) and the HMBC correlation between the methyl protons with 26 (δC 37.3 ppm) and 28 (δC 55.0 ppm) carbons (Figures S1 and S5). The signal at δH 9.46 ppm was assigned to phenylalaninal aldehyde proton. The occurrence of the studied compound in the linear form was further confirmed by the lack of the ROESY correlation between tyrosine and phenylalanine residues.
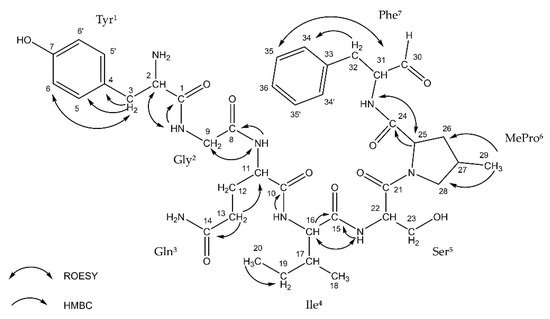
Figure 1. ROESY and HMBC correlations in nostocyclopeptide Ncp-A2-L.
Table 1. Structures of six nostocyclopeptide-variants isolated from Nostoc edaphicum CCNP1411 as pure compounds.
| Peptide Name | Molecular Mass | Structure |
|---|---|---|
| Ncp-A1 | 756 | [Tyr1+Gly2+Gln3+Ile4+Ser5+MePro6+Leu7] |
| Ncp-A2 | 790 | [Tyr1+Gly2+Gln3+Ile4+Ser5+MePro6+Phe7] |
| Ncp-A2-L | 808 | Tyr1+Gly2+Gln3+Ile4+Ser5+MePro6+Phe-H7 |
| Ncp-E2 | 742 | [Tyr1+Gly2+Gln3+Ile4+Ser5+Pro6+Leu7] |
| Ncp-E2-L | 760 | Tyr1+Gly2+Gln3+Ile4+Ser5+Pro6+Leu-H7 |
| Ncp-E4-L | 676 | Tyr1+Gly2+Gln3+Ile4+Ser5+MePro6 |
Table 2. Nuclear Magnetic Resonance (NMR) Spectroscopic Data for Ncp-A2-L (Tyr-Gly-Gln-Ile-Ser-MePro-Phe-H).
| Residue | Position | δC, type | δH (J in Hz) | ROESY | HMBC a |
|---|---|---|---|---|---|
| Tyr | 1 | ||||
| 2 | 169.9, C | ||||
| 3 | 54.6, CH | 4.13, t (6.9, 6.9) | NH(1), 6 | ||
| 4 | 36.0, CH2 | 3.04, dd (7.3, 12.9) | 6 | ||
| 5/5′ | 125.5, C | ||||
| 6/6′ | 130.9, CH | 6.78, d (8.0) | 2, 4, 5 | ||
| 7 | 115.9, CH | 7.04, d (8.0) | 2, 3 | ||
| NH2 | 155.3, C | ||||
| OH | |||||
| Gly | 8 | 170.7, C | |||
| 9 | 42.4, CH2 | 3.84, m | NH(2) | ||
| NH(1) | 8.46, t (5.6, 5.6) | 2 | 1 | ||
| Gln | 10 | ||||
| 11 | 173.1, C | NH(3) | |||
| 12a | 53.2, CH | 4.30, m | 10 | ||
| 12b | 27.2, CH2 | 1.88, m | |||
| 13 | 1.99, m | ||||
| 14 | 31.1, CH2 | 2.26, t (7.3, 7.3) | 11, 12, 14 | ||
| NH(2) | 178.0, C | 9 | |||
| NH2 | 8.25, d (7.6) | 8 | |||
| Ile | 15 | 173.4, C | |||
| 16 | 58.2, CH | 4.09, t (8.1, 8.1) | NH(4) | 17 | |
| 17 | 36.0, CH | 1.77, m | |||
| 18 | 14.7, CH3 | 1.08, d (6.6) | |||
| 19 | 24.6, CH2 | 1.32, m | |||
| 20 | 10.0, CH3 | 0.79, t (7.3, 7.3) | 22 | 17, 19 | |
| NH(3) | 8.21, d (6.8) | 11 | 10 | ||
| Ser | 21 | ||||
| 22 | n.o. | ||||
| 23a | n.o. | 4.59, m | 20 | ||
| 23b | 61.0, CH2 | 3.69, m | |||
| NH(4) | 3.77, m | ||||
| OH | 8.31, d (5.1) | 16 | 15 | ||
| MePro | 24 | 173.8, C | |||
| 25 | 61.3, CH | 4.15, dd (8.1, 9.3) | 24 | ||
| 26 | 37.3, CH2 | 2.13, m | |||
| 27 | 33.1, CH | 2.04, m | |||
| 28a | 55.0, CH2 | 2.84, t (10.5, 10.5) | NH(5) | ||
| 28b | 3.86, m | 26 | |||
| 29 | 15.2, CH3 | 0.82, d (6.6) | 26, 28 | ||
| Phe-H | 30 | 9.46, s | |||
| 31 | n.o. | 4.01, m | 35 | ||
| 32a | 55.5, CH | 2.57, dd (10.6, 14.0) | 34 | 34/34′ | |
| 32b | 34.2, CH2 | 2.98, dd (4.0, 14.5) | |||
| 33 | 32 | ||||
| 34/34′ | 137.9, C | 7.26, m | |||
| 35/35′ | 129.5, CH | 7.15, d (7.2) | 31 | ||
| 36 | 128.7, CH | 7.18, m | |||
| NH(5) | 126.6, CH | 7.46, d (9.3) | 25 | 24 |
a HMBC correlations are given from proton(s) stated to the indicated carbon atom.
In our preliminary studies with the application of the human 20S proteasome, the Ncp-containing fractions of CCNP1411 inhibited CT-L activity at micromolar concentrations. In the current work, to unequivocally state which of the cyanobacterial metabolites are responsible for this activity, the six isolated Ncps were assayed. For three cyclic Ncps (Ncp-A1, Ncp-A2, Ncp-E2) and the six-amino acid linear variant without an aldehyde group (Ncp-E4-L), no effects on CT-L activity of the human 20S proteasome were observed (Figure 2A). This activity was inhibited only by two linear peptide aldehydes, Ncp-A2-L and Ncp-E2-L, applied at 50 µM (Figure 2A). As the two Ncps differ in position 6 (Pro/MePro) and the C-terminal amino acid (Leu/Phe), it can be concluded that these residues do not affect the CT-L activity. Nostocyclopeptide Ncp-E2-L, as well as the widely used synthetic proteasome inhibitor MG-132 [25][26], contain the aldehyde group on C-terminal Leu. The potent activity of MG-132 (IC50 0.11 µM) [27] was attributed to the formation of the hemiacetal covalent bond between the aldehyde group of C-terminal Leu and the hydroxyl group of Thr1 present in the active site of the proteasome [28]. Another bioactive linear nostocyclopeptide from CCNP1411, Ncp-A2-L, also has the C-terminal amino acid aldehyde (Phe), which again confirms the importance of the aldehyde group for the CT-L inhibition [14][29]. Due to the limited amounts of the isolated Ncps, their effects on T-L and C-L activities were examined with no replications. In the assays, only the cyclic Ncp-A2 showed concentration-dependent inhibition of T-L activity (Figure 2B) and had weak effects on C-L activity (Figure 2C). The other Ncp variants had no clear effects on the two proteolytic sites.
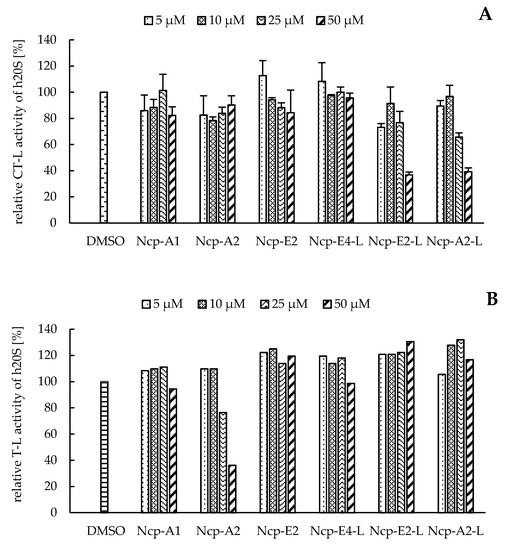
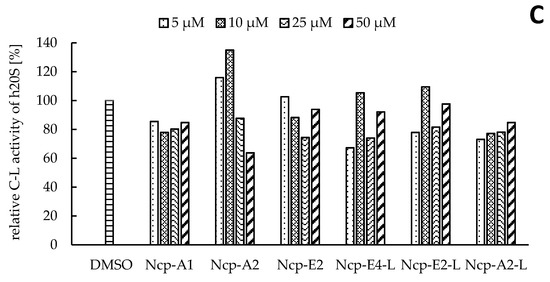
Figure 2. The effects of cyclic and linear (L) nostocyclopeptides (Ncps) on the CT-L (A), T-L (B), and C-L (C) activities of the human 20S proteasome. DMSO was used as a control, and PR11 was used to ensure the correctness of the assay. In the CT-L activity assay, all Ncp variants were tested in triplicate.
The two linear Ncps, Ncp-A2-L and Ncp-E2-L, moderately decreased the CT-L activity (IC50 ca. 50 µM), compared with several known aldehyde-containing proteasome inhibitors [13]. However, this moderate potency of the Ncps is compensated by their high specificity. Unlike many other peptide aldehydes, which inhibit a wide range of proteases [13][29], the two Ncps interacted with the CT-L site but did not modify the T-L and C-L activities.
Among cyanobacteria metabolites, α,β-epoxyketones carmaphycin A and B, isolated from Symploca sp., were found to inhibit CT-L activity of the Saccharomyces cerevisiae 20S proteasome at low nanomolar concentrations [30]. The authors suggested that the sulfoxide/sulfone moieties in the methionine-derived residues of the inhibitor are crucial for the interaction with the enzyme complex. Nostodione A from Scytonema hofmannii, which inhibits CT-L activity (IC50 50 μM), contains indole moiety fused with diketone system [31]. Other cyclic metabolites from cyanobacteria have demonstrated inhibitory effects against 20S complex. For example, scytonemide A from S. hofmannii, a cyclic peptide characterized by the presence of a unique imino linkage, inhibited catalytic activity of proteasome at IC50 96 nM [32]. Krunic et al. [32] suggested that Gln residue contributed to structural conformation in this peptide, enabling optimal binding at the active site. On the other hand the presence of an imine enabled the formation of a covalent bond. Nostoc-derived (Nostoc sp. UIC 10022A) cylindrocyclophanes were active against the 20S proteasome in a wide range of activities (IC50 2.2–100 μM). According to the authors, dichloromethyl moiety was crucial to achieving a higher level of inhibition [33].
The proteasome is an important drug target in a variety of diseases [10][11][12]. Currently, three proteasome inhibitors, approved by the American Food and Drug Administration (FDA), are clinically used for the treatment of multiple myeloma (MM) and mantle cell lymphoma (MVL) patients: bortezomib (Velcade) [34], carfilzomib (Kyprolis) [35] and ixazomib (Ninlaro) [36]. Unfortunately, despite the initial promising effects and high efficacy of the proteasome inhibitors in MM treatment, in many patients, resistance has developed. Moreover, some patients do not respond to this treatment, or the side effects of the drugs are too severe [37][38][39][40].
Currently, the application of proteasome inhibitors in the treatment of other diseases, e.g., in autoimmune disorders, inflammation, or malaria, is explored [4][41]. Further studies are also performed to better understand the effect of specific proteasome inhibitors on general protein homeostasis. In parallel, screening for novel agents with a potential therapeutic application as regulators of the 20S proteasome and other components of the ubiquitin-proteasome system is continued [42][43]. Regardless of the low potency, Ncps still can be considered as starting points for drug development. As in the case of many bioactive natural products, their activity and selectivity can be optimized by structural modifications so that the final compound can demonstrate a better therapeutic potential.
This entry is adapted from the peer-reviewed paper 10.3390/biom11101483
References
- Kisselev, A.F.; Callard, A.; Goldberg, A. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J. Biol. Chem. 2006, 281, 8582–8590.
- Adams, J. The proteasome: Structure, function, and role in the cell. Cancer Treat. Rev. 2003, 29, 3–9.
- Deshmukh, F.K.; Yaffe, D.; Olsshina, M.A.; Ben-Nissan, G.; Sharon, M. The contribution of the 20S proteasome to proteostasis. Biomolecules 2019, 9, 190.
- Sherman, D.J.; Li, J. Proteasome inhibitors: Harnessing proteostasis to combat disease. Molecules 2020, 25, 671.
- Ciechanover, A.; Brundin, P. The ubiquitin proteasome system in neurodegenerative diseases: Sometimes the chicken, sometimes the egg. Neuron 2003, 40, 427–446.
- Ciechanover, A. Proteolysis: From the lysosome to ubiquitin and the proteasome. Nat. Rev. 2005, 6, 79–86.
- Bedford, L.; Paine, S.; Sheppard, P.W.; Mayer, R.J.; Roelofs, J. Assembly, structure and function of the 26S proteasome. Trends Cell Biol. 2010, 20, 391–401.
- Orlowski, M.; Wilk, S. Ubiquitin-independent proteolytic functions of the proteasome. Arch. Biochem. Biophys. 2003, 415, 1–5.
- Hwang, J.; Winkler, L.; Kalejta, R.F. Ubiquitin-independent proteasomal degradation during oncogenic viral infections. Biochim. Biophys. Acta 2011, 1816, 147–157.
- Tundo, G.R.; Sbardella, D.; Santoro, A.M.; Coletta, A.; Oddone, F.; Grasso, G.; Milardi, D.; Lacal, P.M.; Marini, S.; Purrello, L.; et al. The proteasome as a druggable target with multiple therapeutic potentialities: Cutting and non-cutting edges. Pharmacor. Ther. 2020, 213, 107579.
- Verbrugge, S.E.; Scheper, R.J.; Lems, W.F.; de Gruijl, T.D.; Jansen, G. Proteasome inhibitors as experimental therapeutics of autoimmune diseases. Arthritis Res. Ther. 2015, 17, 17.
- Cao, Y.; Zhu, H.; He, R.; Kong, L.; Shao, J.; Zhuang, R.; Xi, J.; Zhang, J. Proteasome, a promising therapeutic target for multiple diseases beyond cancer. Drug Des. Devel. Ther. 2020, 14, 4327–4342.
- de Bettignies, G.; Coux, O. Proteasome inhibitors: Dozens of molecules and still counting. Biochimie 2010, 92, 1530–1545.
- Ma, Y.; Xu, B.; Fang, Y.; Yang, Z.; Cui, J.; Zhang, L.; Zhang, L. Synthesis and SAR study of novel peptide aldehydes as inhibitors of 20S proteasome. Molecules 2011, 16, 7551–7564.
- Harer, S.L.; Bhatia, M.S.; Bhatia, N.M. Proteasome inhibitors mechanism; source for design of newer therapeutic agents. J. Antibiot. 2012, 65, 279–288.
- Oerlemans, R.; Berkers, C.; Assaraf, Y.G.; Scheffer, G.L.; Peters, G.J.; Verbrugge, S.E.; Cloos, J.; Slootstra, J.; Meloen, R.H.; Shoemaker, R.H.; et al. Proteasome inhibition and mechanism of resistance to a synthetic, library-based hexapeptide. Investig. New Drugs 2018, 36, 797–809.
- Momose, I.; Sekizawa, R.; Hashizume, H.; Kinoshita, N.; Homma, Y.; Hamada, M.; Iinuma, H.; Takeuchi, T. Tyropeptins A and B, new proteasome inhibitors produced by Kitasatospora sp. MK993-dF2. J. Antibiot. 2001, 54, 997–1003.
- Momose, I.; Sekizawa, R.; Iinuma, H.; Takeuchi, T. Inhibition of proteasome activity by tyropeptin A in PC12 cells. Biosci. Biotechnol. Biochem. 2002, 66, 2256–2258.
- Hines, J.; Groll, M.; Fahnestock, M.; Crews, C.M. Proteasome inhibition by fellutamide B induces nerve growth factor synthesis. Chem. Biol. 2008, 15, 501–512.
- Golakoti, T.; Yoshida, W.; Chaganty, S.; Moore, R. Isolation and structure determination of nostocyclopeptides A1 and A2 from the terrestrial cyanobacterium Nostoc sp. ATCC53789. J. Nat. Prod. 2001, 64, 54–59.
- Jokela, J.; Herfindal, L.; Wahlsten, M.; Permi, P.; Selheim, F.; Vasconçelos, V.; Døskeland, S.; Sivonen, K. A novel cyanobacterial nostocyklopeptide is a potent antitoxin against Microcystis. ChemBioChem 2010, 11, 1594–1599.
- Nowruzi, B.; Khavari-Nejad, R.; Sivonen, K.; Kazemi, B.; Najafi, F.; Nejadsattari, T. Identification and toxigenic potential of Nostoc sp. Algae 2012, 27, 303–313.
- Liamer, A.; Jensen, J.; Dittman, E. A genetic and chemical perspective on symbiotic recruitment of cyanobacteria of the genus Nostoc into the host plant Blasia pusilla L. Front. Microbiol. 2016, 7, 1963.
- Fidor, A.; Grabski, M.; Gawor, J.; Gromadka, R.; Węgrzyn, G.; Mazur-Marzec, H. Nostoc edaphicum CCNP1411 from the Baltic Sea—a new producer of nostocyclopeptides. Mar. Drugs 2020, 18, 442.
- Guo, N.; Zhilan, P. MG 132, a proteasome inhibitor, induces apoptosis in tumor cells. Asia Pac. J. Clin. Oncol. 2013, 9, 6–11.
- Zhang, L.; Hu, J.J.; Gong, F. MG 132 inhibition of proteasome blocks apoptosis induced by severe DNA damage. Cell Cycle 2011, 10, 3515–3518.
- Hasegawa, M.; Kinoshita, K.; Nishimura, C.; Matsumura, U.; Shionyu, M.; Ikeda, S.; Mizukami, T. Affinity labeling of the proteasome by a belactosin A derived inhibitor. Bioorg. Med. Chem. Lett. 2008, 18, 5668–5671.
- Kisselev, A.F.; van der Linden, W.A.; Overkleeft, H.S. Proteasome inhibitors: An expanding army attacking a unique target. Chem. Biol. 2012, 19, 99–115.
- Kisselev, A.F.; Goldberg, A. Proteasome inhibitors: From research tools to drug candidates. Chem. Biol. 2001, 8, 739–758.
- Pereira, A.R.; Kale, A.J.; Fenley, A.T.; Byrum, T.; Debonsi, H.M.; Gilson, M.K.; Valeriote, F.A.; Moore, B.S.; Gerwick, W.H. The carmaphycins: New proteasome inhibitors exhibiting an α,β-epoxyketone warhead from a marine cyanobacterium. ChemBioChem 2012, 13, 810–817.
- Shim, S.H.; Chlipala, G.; Orjala, J. Isolation and structure determination of a proteasome inhibitory metabolite from a culture of Scytonema hofmanni. J. Microbiol. Biotechnol. 2008, 18, 1655–1658.
- Krunic, A.; Vallat, A.; Mo, S.; Lantvit, D.D.; Swanson, S.M.; Orjala, J. Scytonemides A and B, cyclic peptides with 20S proteasome inhibitory activity from the cultured cyanobacterium Scytonema hofmanii. J. Nat. Prod. 2010, 29, 1927–1932.
- Chlipala, G.E.; Sturdy, M.; Krunic, A.; Lantvit, D.D.; Shen, Q.; Porter, K.; Swanson, S.M.; Orjala, J. Cylindrocyclophanes with proteasome inhibitory activity from the cyanobacterium Nostoc sp. J. Nat. Prod. 2010, 73, 1529–1537.
- Field-Smith, A.; Morgan, G.J.; Davies, F.E. Bortezomib (VelcadeTM) in the treatment of multiple myeloma. Ther. Clin. Risk Manag. 2006, 2, 271–279.
- Herndon, T.M.; Deisseroth, A.; Kaminskas, E.; Kane, R.C.; Koti, K.M.; Rothmann, M.D.; Habtemariam, B.; Bullock, J.; Bray, J.D.; Hawes, J.; et al. Food and Drug Administration approval: Carfilzomib for the treatment of multiple myeloma. Clin. Cancer Res. 2013, 19, 4559–4563.
- Shirley, M. Ixazomib: First global approval. Drugs 2016, 76, 405–411.
- Bai, Y.; Su, X. Updates to the drug-resistant mechanism of proteasome inhibitors in multiple myeloma. Asia-Pac. J. Clin. Oncol. 2021, 17, 29–35.
- Kim, K.B. Proteasomal adaptations to FDA-approved proteasome inhibitors: A potential mechanism for drug resistance. Cancer Drug Resist. 2021, 4, 634–645.
- Moreau, P.; Richardson, P.G.; Cavo, M.; Orlowski, R.Z.; Sam Miguel, J.F.; Palumbo, A.; Harousseau, J.-L. Proteasome inhibitors in multiple myeloma: 10 years later. Blood 2012, 120, 947–959.
- Pancheri, E.; Guglielmi, V.; Wilczynski, G.M.; Malatesta, M.; Tonin, P.; Tomelleri, G.; Nowis, D.; Vattemi, G. Non-hematologic toxicity of bortezomib in multiple myeloma: The neuromuscular and cardiovascular adverse effects. Cancers 2020, 12, 2540.
- Cromm, P.M.; Crews, C.M. The proteasome in modern drug discovery: Second life of highly valuable drug target. ACS Cent. Sci. 2017, 3, 830–838.
- Mofers, A.; Selvaraju, K.; Gubat, J.; D’Arcy, P.; Linder, S. Identification of proteasome inhibitors using analysis of gene. Eur. J. Pharmacol. 2020, 889, 173709.
- Shen, X.; Wu, C.; Lei, M.; Yan, Q.; Zhang, H.; Zhang, L.; Wang, X.; Yang, Y.; Li, J.; Zhu, Y.; et al. Anti-tumor activity of a novel proteasome inhibitor D395 against multiple myeloma and its lower cardiotoxicity compared with carfilzomib. Cell Death Dis. 2021, 12, 429.
This entry is offline, you can click here to edit this entry!
