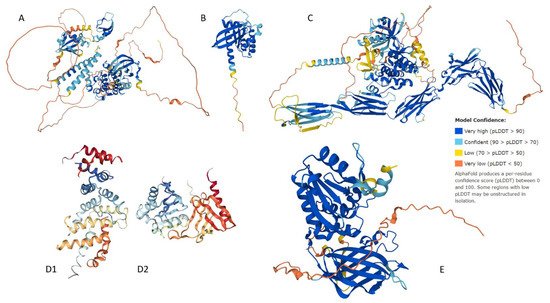
In recent years, there has been tremendous enthusiasm with respect to detailing the genetic basis of many neoplasms, including conjunctival melanoma (CM). We analyze five proteins associated with CM, namely BRAF, NRAS, c-KIT, NF1, and PTEN. We generated structures of BRAF, NRAS, c-KIT, and PTEN with AlphaFold and sourced NF1’s structure from the Protein Databank (PDB). The Predictor of Natural Disordered Protein Regions (PONDR®), the web server for the prediction of intrinsically unstructured regions of proteins (IUPred), and the mean disorder profile (MDP) was utilized to analyze each protein for intrinsically disordered protein regions (IDPRs). AlphaFold and PDB structures show IDPRs in all five proteins. The PONDR®, IUPred, and MDP analysis confirmed high levels of IDPRs. By PONDR®, the highest levels of IDPRs were in BRAF (45.95%), followed by PTEN (31.76%), NF1 (22.19%), c-KIT (21.82%), and NRAS (14.81%). Our STRING analysis found that each of these five proteins had more predicted interactions then expected (p-value < 1.0 × 10−16). Our analysis demonstrates that the mutations linked to CM likely affected IDPRs and possibly altered their highly complex PPIs. Quantifying IDPRs in BRAF, NRAS, c-KIT, NF1, and PTEN and understanding these protein regions are important processes as IDPRs can be possible drug targets for novel targeted therapies for treating CM.
- conjunctival melanoma
- genetics
- structural analysis
- intrinsically disordered proteins
- targeted therapies
- mitogen-activated protein kinase (MAPK)
- Phosphatidylinositol-3-Kinase and Protein Kinase B (PI3K-akt)
- proto-oncogene B-Raf (BRAF)
- neuroblastoma v-ras o
1. Structural Assessment
Table 1. Genes with mutations known to be associated with the development of conjunctival melanoma.
| Gene Name | Know Gene Mutation | UniProt ID | Structure Source |
| BRAF | G469A, D594G, V600E/K/R | P15056 | AlphaFold2 |
| NRAS | Q12C, Q13D, Q61R/H/L/K | P01111 | AlphaFold2 |
| c-KIT | * | P10721 | AlphaFold2 |
| NF1 | More then 25 distinct mutations | P21359 | 1NF1and 3PEG (PDB IDs) |
| PTEN | Chromosome 10q deletion | P60484 | AlphaFold2 |
2. Quantitative Disorder Based Predictions
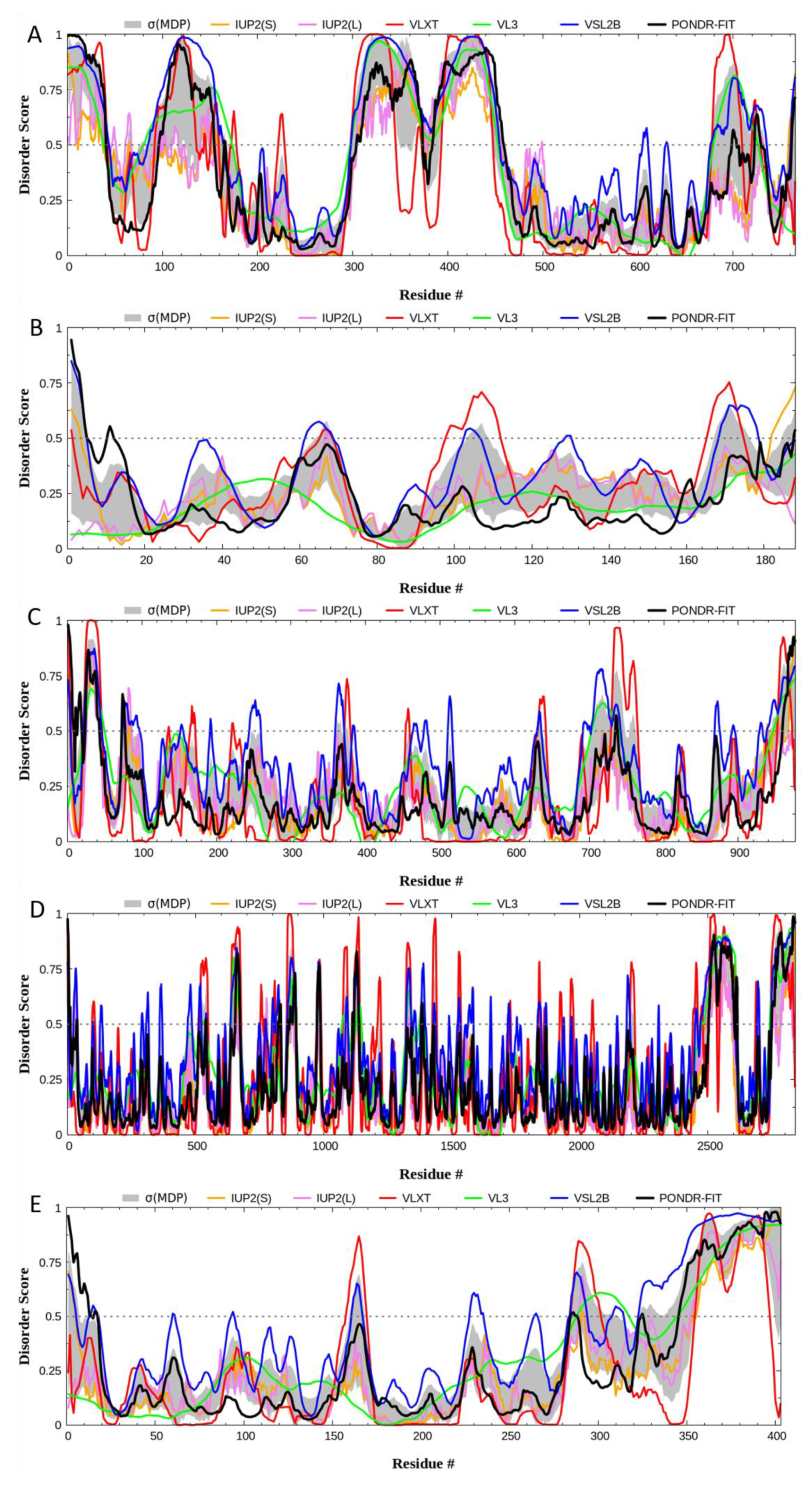
|
|
Gene Name |
||||
|
Predictor |
BRAF |
NRAS |
c-KIT |
NF1 |
PTEN |
|
PONDR® VLXT |
33.55% | 14.29% | 12.30% | 19.90% | 17.62% |
|
PONDR® VL3 |
43.08% | 0.00% | 9.53% | 13.46% | 22.08% |
|
PONDR® VSL2 |
45.95% | 14.81% | 21.82% | 22.19% | 31.76% |
|
Average |
40.86% | 9.70% | 14.55% | 18.52% | 23.82% |
3. Protein-Protein Interaction Network
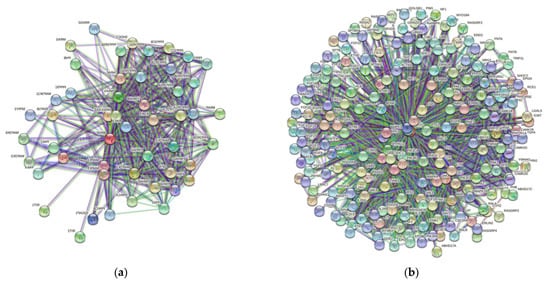
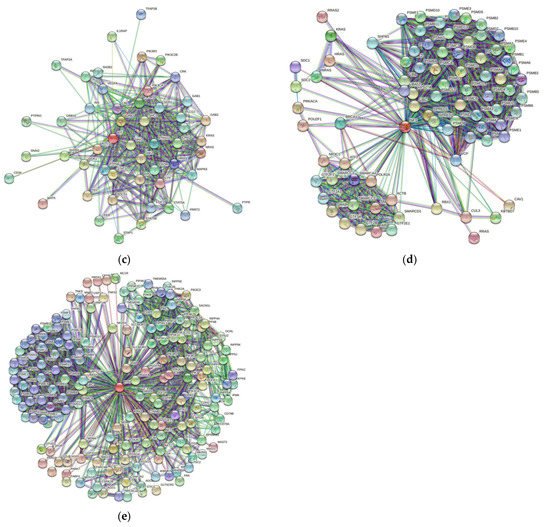
| Gene Name | Proteins in Network | Expected Number of Interactions | Predicted Number of Interactions | p-Value |
|---|---|---|---|---|
| BRAF | 109 | 253 | 2213 | <10−16 |
| NRAS | 327 | 1619 | 7795 | <10−16 |
| c-KIT | 60 | 177 | 495 | <10−16 |
| NF1 | 90 | 388 | 1790 | <10−16 |
| PTEN | 157 | 622 | 2297 | <10−16 |
3. Discussion
This entry is adapted from the peer-reviewed paper 10.3390/genes12101625
References
- Gargi K. Vora; Hakan Demirci; Brian Marr; Prithvi Mruthyunjaya; Advances in the management of conjunctival melanoma. Survey of Ophthalmology 2017, 62, 26-42, 10.1016/j.survophthal.2016.06.001.
- UniProt Consortium; UniProt: a worldwide hub of protein knowledge . Nucleic Acids Research 2019, 47, D506-D515, .
- John Jumper; Richard Evans; Alexander Pritzel; Tim Green; Michael Figurnov; Olaf Ronneberger; Kathryn Tunyasuvunakool; Russ Bates; Augustin Žídek; Anna Potapenko; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583-589, 10.1038/s41586-021-03819-2.
- Meszaros, Balint; Edros, Gabor; Dosztanyi, Zsuzsanna; IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding . Nucleic Acid Research 2018, 46, W329-W337, .
- Pedro Romero; Zoran Obradovic; Xiaohong Li; Ethan C. Garner; Celeste J. Brown; A. Keith Dunker; Sequence complexity of disordered protein. Proteins: Structure, Function, and Genetics 2000, 42, 38-48, 10.1002/1097-0134(20010101)42:1%3c38::AID-PROT50%3e3.0.CO;2-3.
- Kang Peng; Slobodan Vucetic; Predrag Radivojac; Celeste J. Brown; A. Keith Dunker; Zoran Obradovic; OPTIMIZING LONG INTRINSIC DISORDER PREDICTORS WITH PROTEIN EVOLUTIONARY INFORMATION. Journal of Bioinformatics and Computational Biology 2005, 3, 35-60, 10.1142/s0219720005000886.
- Bin Xue; Roland Dunbrack; Robert W. Williams; A. Keith Dunker; Vladimir N. Uversky; PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 2010, 1804, 996-1010, 10.1016/j.bbapap.2010.01.011.
- Krithika Rajagopalan; Steven M. Mooney; Nehal Parekh; Robert H. Getzenberg; Prakash Kulkarni; A majority of the cancer/testis antigens are intrinsically disordered proteins. Journal of Cellular Biochemistry 2011, 112, 3256-3267, 10.1002/jcb.23252.
- Anastasia Gkiala; Sotiria Palioura; Conjunctival Melanoma: Update on Genetics, Epigenetics and Targeted Molecular and Immune-Based Therapies. Clinical Ophthalmology 2020, 14, 3137-3152, 10.2147/opth.s271569.
- Mateusz Biesaga; Marta Frigolé-Vivas; Xavier Salvatella; Intrinsically disordered proteins and biomolecular condensates as drug targets. Current Opinion in Chemical Biology 2021, 62, 90-100, 10.1016/j.cbpa.2021.02.009.
- Patricia Santofimia-Castaño; Bruno Rizzuti; Yi Xia; Olga Abian; Ling Peng; Adrian Velazquez-Campoy; José L. Neira; Juan Iovanna; Targeting intrinsically disordered proteins involved in cancer. Experientia 2020, 77, 1695-1707, 10.1007/s00018-019-03347-3.
- Filippo Spriano; Elaine Yee Lin Chung; Eugenio Gaudio; Chiara Tarantelli; Luciano Cascione; Sara Napoli; Katti Jessen; Laura Carrassa; Valdemar Priebe; Giulio Sartori; et al. The ETS Inhibitors YK-4-279 and TK-216 Are Novel Antilymphoma Agents. Clinical Cancer Research 2019, 25, 5167-5176, 10.1158/1078-0432.ccr-18-2718.
- Marianne D. Sadar; Discovery of drugs that directly target the intrinsically disordered region of the androgen receptor. Expert Opinion on Drug Discovery 2020, 15, 551-560, 10.1080/17460441.2020.1732920.
- José L. Neira; Jennifer Bintz; María Arruebo; Bruno Rizzuti; Thomas Bonacci; Sonia Vega; Angel Lanas; Adrián Velázquez-Campoy; Juan L. Iovanna; Olga Abián; et al. Identification of a Drug Targeting an Intrinsically Disordered Protein Involved in Pancreatic Adenocarcinoma. Scientific Reports 2017, 7, 39732, 10.1038/srep39732.
- Pietro Quaglino; Paolo Fava; Luca Tonella; Marco Rubatto; Simone Ribero; Maria Teresa Ferro; Treatment of Advanced Metastatic Melanoma. Dermatology Practical & Conceptual 2021, 11, 2021164S-2021164S, 10.5826/dpc.11s1a164s.
- Bin Wu; Lizheng Shi; FrontlineBRAFTesting–Guided Treatment for Advanced Melanoma in the Era of Immunotherapies. JAMA Dermatology 2020, 156, 1177, 10.1001/jamadermatol.2020.2398.
- Bin Xue; Roland Dunbrack; Robert W. Williams; A. Keith Dunker; Vladimir N. Uversky; PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 2010, 1804, 996-1010, 10.1016/j.bbapap.2010.01.011.
- Krithika Rajagopalan; Steven M. Mooney; Nehal Parekh; Robert H. Getzenberg; Prakash Kulkarni; A majority of the cancer/testis antigens are intrinsically disordered proteins. Journal of Cellular Biochemistry 2011, 112, 3256-3267, 10.1002/jcb.23252.
