Coronavirus disease 19 (COVID-19) is caused by an enveloped, positive-sense, single-stranded RNA virus, referred to as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which belongs to the realm Riboviria, order Nidovirales, family Coronaviridae, genus Betacoronavirus and the species Severe acute respiratory syndrome-related coronavirus. This viral disease is characterized by a myriad of varying symptoms, such as pyrexia, cough, hemoptysis, dyspnoea, diarrhea, muscle soreness, dysosmia, lymphopenia and dysgeusia amongst others. The virus mainly infects humans, various other mammals, avian species and some other companion livestock. SARS-CoV-2 cellular entry is primarily accomplished by molecular interaction between the virus’s spike (S) protein and the host cell surface receptor, angiotensin-converting enzyme 2 (ACE2), although other host cell-associated receptors/factors, such as neuropilin 1 (NRP-1) and neuropilin 2 (NRP-2), C-type lectin receptors (CLRs), as well as proteases such as TMPRSS2 (transmembrane serine protease 2) and furin, might also play a crucial role in infection, tropism, pathogenesis and clinical outcome.
- SARS-CoV-2
- coronavirus disease 19
- pathogenesis
- therapeutic targeting
1. Potential Therapeutic Targets on Host Cells
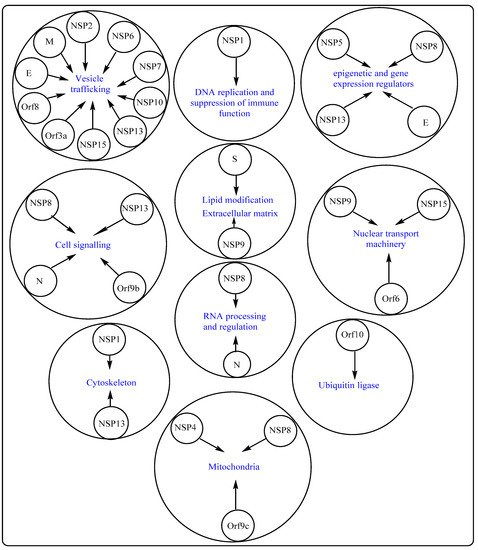
2. Potential Therapeutic Targets on SARS-CoV-2
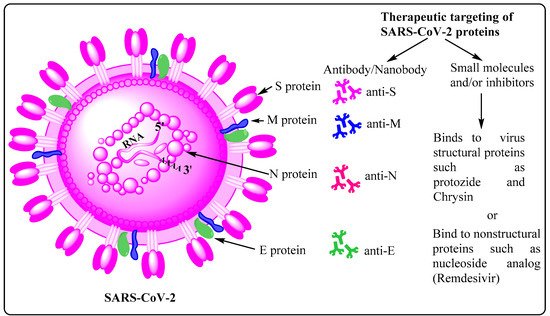
This entry is adapted from the peer-reviewed paper 10.3390/cells10092427
References
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.
- Day, C.J.; Bailly, B.; Guillon, P.; Dirr, L.; Jen, F.E.-C.; Spillings, B.L.; Mak, J.; von Itzstein, M.; Haselhorst, T.; Jennings, M.P. Multidisciplinary Approaches Identify Compounds that Bind to Human ACE2 or SARS-CoV-2 Spike Protein as Candidates to Block SARS-CoV-2–ACE2 Receptor Interactions. mBio 2021, 12.
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.
- Zang, R.; Castro, M.F.G.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B.; et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, eabc3582.
- Singh, M.; Bansal, V.; Feschotte, C. A Single-Cell RNA Expression Map of Human Coronavirus Entry Factors. Cell Rep. 2020, 32, 108175.
- Lambert, S.; Bouttier, M.; Vassy, R.; Seigneuret, M.; Petrow-Sadowski, C.; Janvier, S.; Heveker, N.; Ruscetti, F.W.; Perret, G.; Jones, K.S.; et al. HTLV-1 uses HSPG and neuropilin-1 for entry by molecular mimicry of VEGF165. Blood 2009, 113, 5176–5185.
- Wang, H.-B.; Zhang, H.; Zhang, J.-P.; Li, Y.; Zhao, B.; Feng, G.-K.; Du, Y.; Xiong, D.; Zhong, Q.; Liu, W.-L.; et al. Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat. Commun. 2015, 6, 6240.
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; Van Der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860.
- Guo, H.-F.; Kooi, C.W.V. Neuropilin Functions as an Essential Cell Surface Receptor. J. Biol. Chem. 2015, 290, 29120–29126.
- Llorián-Salvador, M.; González-Rodríguez, S. Painful Understanding of VEGF. Front. Pharm. 2018, 9, 1267.
- Jarvis, A.; Allerston, C.K.; Jia, H.; Herzog, B.; Garza-Garcia, A.; Winfield, N.; Ellard, K.; Aqil, R.; Lynch, R.; Chapman, C.; et al. Small Molecule Inhibitors of the Neuropilin-1 Vascular Endothelial Growth Factor A (VEGF-A) Interaction. J. Med. Chem. 2010, 53, 2215–2226.
- Wei, J.; Alfajaro, M.M.; DeWeirdt, P.C.; Hanna, R.E.; Lu-Culligan, W.J.; Cai, W.L.; Strine, M.S.; Zhang, S.-M.; Graziano, V.R.; Schmitz, C.O.; et al. Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell 2020, 184, 76–91.
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468.
- Kondo, T.; Watanabe, M.; Hatakeyama, S. TRIM59 interacts with ECSIT and negatively regulates NF-κB and IRF-3/7-mediated signal pathways. Biochem. Biophys. Res. Commun. 2012, 422, 501–507.
- Li, S.; Wang, L.; Berman, M.; Kong, Y.-Y.; Dorf, M.E. Mapping a Dynamic Innate Immunity Protein Interaction Network Regulating Type I Interferon Production. Immunity 2011, 35, 426–440.
- Faria, P.A.; Chakraborty, P.; Levay, A.; Barber, G.N.; Ezelle, H.J.; Enninga, J.; Arana, C.; van Deursen, J.; Fontoura, B.M. VSV Disrupts the Rae1/mrnp41 mRNA Nuclear Export Pathway. Mol. Cell 2005, 17, 93–102.
- Nakagawa, K.; Lokugamage, K.; Makino, S. Viral and Cellular mRNA Translation in Coronavirus-Infected Cells. Adv. Virus Res. 2016, 96, 165–192.
- Knoops, K.; Kikkert, M.; Worm, S.H.E.V.D.; Zevenhoven-Dobbe, J.C.; Van Der Meer, Y.; Koster, A.; Mommaas, A.M.; Snijder, E.J. SARS-Coronavirus Replication Is Supported by a Reticulovesicular Network of Modified Endoplasmic Reticulum. PLoS Biol. 2008, 6, e226.
- Shah, P.; Link, N.; Jang, G.M.; Sharp, P.P.; Zhu, T.; Swaney, D.L.; Johnson, J.; Von Dollen, J.; Ramage, H.R.; Satkamp, L.; et al. Comparative Flavivirus-Host Protein Interaction Mapping Reveals Mechanisms of Dengue and Zika Virus Pathogenesis. Cell 2018, 175, 1931–1945.
- Heaton, N.S.; Moshkina, N.; Fenouil, R.; Gardner, T.; Aguirre, S.; Shah, P.; Zhao, N.; Manganaro, L.; Hultquist, J.; Noel, J.; et al. Targeting Viral Proteostasis Limits Influenza Virus, HIV, and Dengue Virus Infection. Immunity 2016, 44, 46–58.
- Benton, D.J.; Wrobel, A.G.; Xu, P.; Roustan, C.; Martin, S.R.; Rosenthal, P.B.; Skehel, J.J.; Gamblin, S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 2020, 588, 327–330.
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Genet. 2020, 19, 155–170.
- Andreano, E.; Nicastri, E.; Paciello, I.; Pileri, P.; Manganaro, N.; Piccini, G.; Manenti, A.; Pantano, E.; Kabanova, A.; Troisi, M.; et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell 2021, 184, 1821–1835.e16.
- Hansen, J.; Baum, A.; Pascal, K.E.; Russo, V.; Giordano, S.; Wloga, E.; Fulton, B.O.; Yan, Y.; Koon, K.; Patel, K.; et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 2020, 369, 1010–1014.
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.-W.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456.
- Baum, A.; Fulton, B.O.; Wloga, E.; Copin, R.; Pascal, K.E.; Russo, V.; Giordano, S.; Lanza, K.; Negron, N.; Ni, M.; et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020, 369, 1014–1018.
- Lytton, S.; Yeasmin, M.; Ghosh, A.; Bulbul, R.H.; Molla, M.A.; Herr, M.; Duchmann, H.; Sharif, M.; Nafisa, T.; Amin, R.; et al. Detection of Anti-Nucleocapsid Antibody in COVID-19 Patients in Bangladesh Is not Correlated with Previous Dengue Infection. Pathogens 2021, 10, 637.
- Heiser, K.; McLean, P.F.; Davis, C.T.; Fogelson, B.; Gordon, H.B.; Jacobson, P.; Hurst, B.; Miller, B.; Alfa, R.W.; Earnshaw, B.A.; et al. Identification of potential treatments for COVID-19 through artificial intelligence-enabled phenomic analysis of human cells infected with SARS-CoV-2. bioRxiv 2020, 054387.
- Yousefi, H.; Mashouri, L.; Okpechi, S.C.; Alahari, N.; Alahari, S.K. Repurposing existing drugs for the treatment of COVID-19/SARS-CoV-2 infection: A review describing drug mechanisms of action. Biochem. Pharm. 2020, 183, 114296.
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293.
- Eastman, R.T.; Roth, J.S.; Brimacombe, K.R.; Simeonov, A.; Shen, M.; Patnaik, S.; Hall, M.D. Correction to Remdesivir: A Review of Its Discovery and Development Leading to Human Clinical Trials for Treatment of COVID-19. ACS Cent. Sci. 2020, 6, 1009.
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271.
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797.
- Di Micco, S.; Musella, S.; Scala, M.C.; Sala, M.; Campiglia, P.; Bifulco, G.; Fasano, A. In silico Analysis Revealed Potential Anti-SARS-CoV-2 Main Protease Activity by the Zonulin Inhibitor Larazotide Acetate. Front. Chem. 2021, 8, 628609.
- Yadav, R.; Hasan, S.; Mahato, S.; Celik, I.; Mary, Y.; Kumar, A.; Dhamija, P.; Sharma, A.; Choudhary, N.; Chaudhary, P.K.; et al. Molecular docking, DFT analysis, and dynamics simulation of natural bioactive compounds targeting ACE2 and TMPRSS2 dual binding sites of spike protein of SARS CoV-2. J. Mol. Liq. 2021, 116942.
- Yadav, R.; Imran, M.; Dhamija, P.; Suchal, K.; Handu, S. Virtual screening and dynamics of potential inhibitors targeting RNA binding domain of nucleocapsid phosphoprotein from SARS-CoV-2. J. Biomol. Struct. Dyn. 2021, 12, 4433–4448.
- Yadav, R.; Imran, M.; Dhamija, P.; Chaurasia, D.K.; Handu, S. Virtual screening, ADMET prediction and dynamics simulation of potential compounds targeting the main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020, 1–16.
- Yadav, R.; Parihar, R.D.; Dhiman, U.; Dhamija, P.; Upadhyay, S.K.; Imran, M.; Behera, S.K.; Prasad, T.S.K. Docking of FDA Approved Drugs Targeting NSP-16, N-Protein and Main Protease of SARS-CoV-2 as Dual Inhibitors. Biointerface Res. Appl. Chem. 2020, 11, 9848–9861.
- Azam, F.; Eid, E.E.M.; Almutairi, A. Targeting SARS-CoV-2 main protease by teicoplanin: A mechanistic insight by docking, MM/GBSA and molecular dynamics simulation. J. Mol. Struct. 2021, 131124.
