The FTO (FaT mass and Obesity-associated) protein is an alpha-ketoglutarate and iron dependent dioxygenase, a member of ALKBH family proteins. FTO removes the methyl groups from modified nucleotides on single stranded DNA or RNA with N6-methyladenozine in the mRNA removed most efficiently. It is engaged in wide range of key physiological processes such as adipogenesis, cell cycle progression, heart remodelling, neural development and osteogenesis. Impairments of FTO activity is lethal or leads to serious developmental disorders. It is also one of the factors responsible for development and maintenance of many type of cancers.
FTO acts in cooperation with other proteins e.g. CaMKII, MRS, SFPQ or XPO2. Similarly to other dioxygenases, it shows ability to form homodimer. Recently, it was shown that FTO interacts with the calmodulin (CaM) in Ca2+ dependent manner.
1. Introduction
The FTO (FaT mass and Obesity-associated) protein is an alpha-ketoglutarate and iron-dependent dioxygenase
[1]. This two-domain protein is responsible for the removal of methyl groups from certain types of RNA
[2]. FTO has been shown to be able to demethylate
N3-methyluridine from RNA oligomers
[3],
N6-methyladenosine (
N6-meA) from mRNA and snRNA,
N1-methyladenosine from tRNA
[4],
N6,2-O-dimethyladenosine from both snRNA
[4] and, the most efficiently, from the mRNA cap
[5]. Importantly, the substrate specificity of FTO may be dependent on its localization within the cell
[4]. The cap demethylation takes place in the cytoplasm only, snRNA demethylation occurs exclusively in the nucleus while the removal of the methyl group from mRNA happens regardless of its location
[4].
The cellular FTO localization depends on its interaction with other proteins. It has been shown that FTO molecules actively move between the nucleus and the cytoplasm. The phosphorylation state of FTO in the position T150, introduced by casein kinase II, determines FTO presence in a particular cell compartment
[6]. Phosphorylation of T150 maintains cytoplasmic location of FTO, while non-phosphorylated molecules are directed to the nucleus. Exportin-2 (XPO2) is one of the proteins involved in FTO transport through recognition of the NLS sequence present at the start of the FTO N-domain
[7].
In the last 10 years, a number of other interactions influencing FTO parameters and regulating its functions in the cell have been identified. Lin and colleagues
[8] have shown that FTO can interact with three (α, β, and γ) out of the four isoforms of calcium/calmodulin-dependent protein kinase II (CaMKII)
[9], the members of the serine/threonine kinase family. CaMKII kinases phosphorylate a wide range of different substrates influencing processes such as cell development, proliferation, cellular transport, neuronal function and regulation of liver glucose
[10][11][12][13][14][15]. For example, the interaction between FTO and CaMKII isoforms decreased the level of cAMP response element-binding protein (CREB) phosphorylation in human neuroblastoma cells (SK-N-SH). Next, the phosphorylation status of CREB protein determined the expression of specific genes associated with regulatory elements CRE
[16]. The observed phosphorylation increased the expression of Neuropeptide Y receptor type 1 (NPY1R) and brain-derived neurotrophic factor (BDNF) proteins, which are also related to appetite and energy homeostasis.
Another protein with confirmed FTO interaction is the splicing factor, proline- and glutamine-rich (SFPQ). This nuclear protein
[17] has been classified as a transcriptional factor participating in several metabolic pathways, e.g., in RNA transport, apoptosis or RNA repair. Together with Non-POU domain-containing octamer-binding proteins (p54nrb) and Paraspeckle component 1 (PSPC1), it forms a multifunctional family of
Drosophila behavior/human splicing (DBHS) proteins with a tendency to create homo- and heterodimers
[17]. Song and colleagues
[18] noted that there are many physiological pathways and pathological conditions with participation of FTO and SFPQ proteins. They found that SFPQ, the RNA-binding protein, interacts with the FTO C-domain
[18] and, when RNA was inspected, both proteins were close to each other on the chain. Moreover, SFPQ protein recognizes the CUGUG sequence and promotes FTO-directed
N6-meA demethylation close to the indicated sequence. It is likely that the regulation of
N6-meA in physiological and pathological states depends on SFPQ-FTO interaction. Additionally, FTO interaction with methionyl-tRNA synthetase (MRS) links the protein with energy metabolism
[19]. MRS is responsible for connection methionine with the corresponding
t-RNA; thus, MRS-FTO interaction suggests FTO involvement in amino acid metabolism.
Phosphorylation of certain serines in FTO by Glycogen synthase kinase 3 (GSK-3) affects the lifetime of the protein by its further ubiquitination, resulting in proteolytic decomposition
[20]. On the other hand, Tai and colleagues
[21] have shown that threonine phosphorylation in FTO gives the opposite effect.
The above-mentioned reports suggest that FTO appears in a network of interactions with other cellular elements. Further, our analysis of protein lysates from head and neck cancer (HNSCC), with the use of size exclusion chromatography indicated that FTO was located in a wide spectrum of molecular fractions—from several tens to several hundred kDa. This confirmed the presence of a wide range of the possible FTO interactors and determined the goal of our present study. Initially, we checked whether potential FTO interactors involve proteins that are known to recognize specific amino acid sequences. The Calmodulin Target Database
[22] enabled us to identify sequences from several species indicating that FTO C-domain potentially interacts with calmodulin (CaM), an important signaling protein. It consists of two globular lobes, N- and C-, each possessing two calcium-binding sites connected through a flexible linker
[23]. We investigated the FTO–CaM relationship, because calmodulin is one of the major regulators of Ca
2+-dependent signaling pathways in all eukaryotic cells. Its high conservation during evolution, broad spectrum of functions via interactions with several dozen of other proteins and the fact that its presence is crucial for many tested organisms
[24] underscore the importance of the research into the interplay between CaM and its partners.
2. FTO Protein Forms Homo- and Heterocomplexes In Vivo
We have already shown that overexpression of the FTO protein occurs in head and neck cancer (HNSCC) and that FTO level was positively correlated with the tumor size, indicating the involvement of this protein in cancer metabolism
[25]. This raises the question whether FTO interactions with various proteins, already reported for CaMKII
[8], GSK-3
[20] or SFPQ
[18], explain its engagement in the metabolic processes other than cancerogenesis, such as adipogenesis
[4], osteogenesis
[26] or neural development
[27] in normal cells. To answer the question of whether regulation of FTOs occurs at the gene or protein level, it must be first determined if in vivo FTO exists as monomer, dimer or in protein complexes of higher molecular mass.
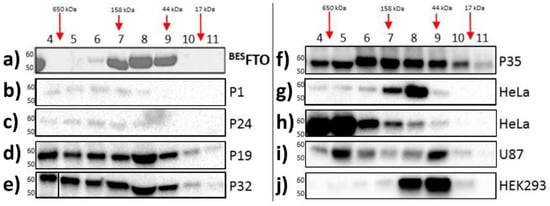
Here, size exclusion chromatography (SEC) was performed for the purified recombinant FTO obtained from the baculovirus expression system (
BESFTO), as well as for samples from the HNSCC tissues and specific cell lines, both cancerous (HeLa and U87) and non-cancerous (HEK293). In the absence of other proteins, FTO (58 kDa) showed the ability to form dimers
[28]. This form predominates in vitro outside the cell. There were no higher oligomeric forms of the protein, even at high concentrations of
BESFTO, that were clearly visible in the Coommasie stained SDS-PAGE gel (
Figure 1a). On the other hand, FTO derived from cellular lysates behaved differently. Western blot analysis of fractions eluted from the column showed that FTO appeared in almost all the fractions, suggesting that in cancer tissues and cancer cell lines the protein exists in highly diverse forms, ranging from monomers up to large complexes (
Figure 1b–j). Interestingly, in human embryonic kidney cells (HEK293), under the tested conditions, FTO appeared rather as a mixture of dimers and predominating monomers. This behavior strongly suggests that FTO may interact with other proteins, which is clearly evidenced by changes in the elution profile. Moreover, the data obtained in the HEK293 cell line suggest that other proteins or small compounds may interfere with FTO homodimerization.
Figure 1. Analysis of cell lysates containing FTO with the use of gel filtration chromatography. Each panel shows FTO contents in particular fractions (4–11) after SEC of protein lysates. (
a) recombinant
BESFTO exists as the mixture of the monomeric and dimeric forms; (
b–
f) HNSCC samples where FTO is present also in high molecular weight fractions, corresponding to masses up to 650 kDa; (
g,
h) HeLa cell samples where FTO is present in different fractions depending on the samples; (
i) U87 cell samples where FTO exist simultaneously in low and high molecular weight fractions; (
j) HEK293 samples where FTO monomeric forms are dominant. The results were visualized by Coomassie staining (
a) or Western blot analysis (
b–
j). Antibodies used in experiment were verified previously
[25].
3. FTO Interacts with CaM via C-Terminal Domain
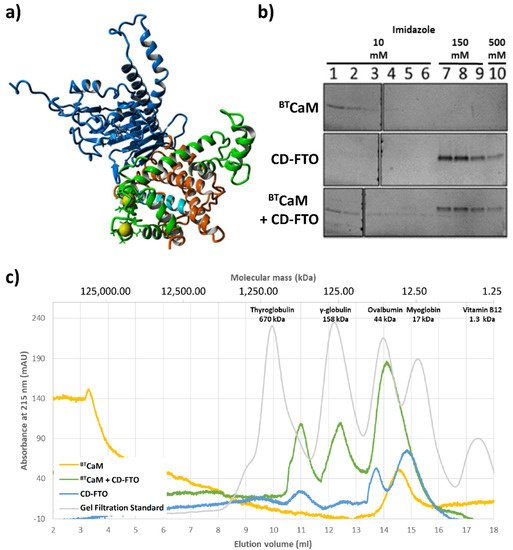
A model of the FTO–CaM complex based on the obtained results is presented on Figure 6a. The N-terminal pair of EF-hand motifs of CaM was found to interact efficiently with FTO, while the C-terminal was oriented unfavorably relative to FTO. In order to avoid the steric hindrances between the C-terminal CaM domain and FTO, various conformations of the CaM interdomain linker were tested, while keeping coordinates of the N-terminal pair of EF-hands fixed. The best model was obtained for the linker in a helical conformation, adopted from the holo form of CaM (1CLL), with the C-terminal domain of CaM remaining calcium free. This model is consistent with the experimental results indicating that the presence of calcium is crucial for interaction and that half of the calmodulin displays different properties than the other half.
Figure 2. In silico and in vitro study of the interaction between CaM and C-domain of ECFTO (CD–FTO). (a) The model of the FTO–CaM–Ca2+ complex. Two calcium (yellow)-binding loops of the N-terminal part of CaM (green) are close to the CaM–biding region (cyan) located in the C-terminal domain (orange) of the FTO (blue). (b,c) Interaction between the ECFTO C–terminal domain with calmodulin. (b) Pull-down assay of BTCaM with the CD–FTO. CD–FTO was used as bait, with selected concentrations of imidazole: 10 mM, 150 mM, 0.5 M. Numbers indicate specific elution fraction in the presence of Fe2+, Ca2+, 2–OG. The presence of the CD–FTO protein in the incubation mixture delayed the elution of BTCaM. Elution experiments with BTCaM only or just CD–FTO were performed as controls. BTCaM alone did not interact with Ni+-resin. (c) Gel filtration chromatography of BTCaM (yellow), CD–FTO (blue) and their mixture (green) samples in the presence of 0.5 mM Mn2+, 0.5 mM Ca2+ and 1 mM 2–OG. The CD–FTO chromatogram shows the presence of two fractions corresponding to monomeric and dimeric state, as well as large unspecific aggregates. BTCaM is present only in one, low molecular mass form. The chromatography of the two protein mixtures reveals the presence of one significant peak corresponding to the expected mass of the CD–FTO–BTCaM complex.
After detailed investigation of CaM behavior in the presence of the FTO, we experimentally verified FTO C-domain interaction with calmodulin. First, the interaction between bovine testicle calmodulin (BTCaM) and FTO C-domain (hereafter called CD-FTO) expressed in the E. Coli system was tested by a pull-down assay where the presence of His-tagged CD-FTO delayed the removal of BTCaM from nickel beads (Figure 2b). Next, complex formation between these proteins was monitored by gel filtration (Figure 2c). The CD-FTO (22.2 kDa) elution profile showed three clearly visible peaks corresponding to large aggregates (at 11.0 mL), a homodimer (at 13.8 mL), and monomer (at 14.9 mL). BTCaM (17 kDa) was eluted as the singular peak at 14.5 mL. As indicated by its molecular mass, it should have eluted at 15.2 mL, similarly to the myoglobin, but because of its non-globular shape, BTCaM was eluted earlier. After gel filtration of the CD-FTO–BTCaM mixture, the elution profile showed three distinct peaks. The first likely corresponds to large aggregates (at 11.0 mL), it was also present in the CD-FTO elution profile. The second (at 12.4 mL) may result from the initial stages of protein aggregation. The CD-FTO–BTCaM mixture was eluted mainly as a peak at 14.1 mL, with the location corresponding to a putative heterodimer. The fact that this peak eluted between the two latter peaks of pure CD-FTO (13.8 mL for the homodimer and 14.9 mL for the monomer) is consistent with the lower molecular mass of BTCaM compared with CD-FTO (17 kDa vs. 22.2 kDa).
4. The importance of FTO-CaM interaction for metabolism
The FTO demethylase is a member of the ALKBH family of proteins connected with both adipogenesis, osteogenesis, maintaining bone mass and heart regeneration, as well as civilization diseases such as obesity, type 2 diabetes, cancer and others
[28]. Many studies have demonstrated that FTO is involved in crucial cell processes; however, not much is known regarding the regulation of its activity. Here, we found that FTO–CaM protein–protein interaction plays a key role in understanding the role of FTO in cellular homeostasis. The amino acid sequence of FTO comprises a fairly conserved motif predicted to be recognized by CaM. Using several complementary approaches, we verified that FTO can indeed interact with CaM at nanomolar concentrations in the presence of Ca
2+. Moreover, the interplay between FTO and CaM was only slightly affected by the type of the expression system used and only in the part of the experiments. Thus, the presence of posttranslational modifications, especially serine phosphorylation
[28], occurring mostly in the FTO N-terminal domain did not significantly affect FTO–CaM binding. The obtained results showing nanomolar
KD are likely to be biologically relevant, because FTO in tissues occurs at nanomolar—or even lower—concentrations that can be detected by sensitive methods, such as Western blot
[29]. On the other hand, total CaM concentration can be much higher. In certain states, it can reach several µM
[30][31] or even 40 µM
[32]. However, the free pool of CaM in a cell may be much lower due to the interactions of CaM with other protein targets, e.g., GAP-43 or RC3
[33][34]. However, it is important to note that the CaM/Ca
2+ affinity to FTO is stronger than many other CaM interactors measured by similar biophysical methods. Here, we estimated
KD for CaM/Ca
2+/FTO complexes to equal ca. 10–30 nM. In contrast, for the CaM/Ca
2+/CaMKII-specific peptide complex (residue 290–309), the
KD measured by MST technique was much higher (190 nM). Meanwhile, the neurogranin protein binds the apo form of calmodulin with even lower affinity:
KD = 890 nM
[35]. Finally, another newly identified CaM interactor, phosphoinositide-interacting regulator of transient receptor potential (PIRT), shows Ca
2+-dependent affinity with
KD of 350 nM and 60 µM in the absence and presence of Ca
2+, respectively
[36].
Having confirmed FTO–CaM interaction, we then investigated the one binding site mode, in comparison with other known calmodulin interactors. According to the Calmodulin Target Database, 350 protein sequences have been shown to bind CaM. The process of CaM-targeted amino acid sequence binding is known to occur both in the presence, as well as in the absence, of Ca2+.
Earlier, Nissen and co-workers
[37] analyzed a number of CaM-targeted complexes and stored them in the Protein Data Bank (PDB). Unsurprisingly, the presence of four Ca
2+ within the CaM-targeted complex was found in the majority of studied PDB structures, because CaM is known to be able to interact with the target proteins/peptides upon binding four Ca
2+. Multiple examples of four Ca
2+-bound CaM-targeted complexes have been reported so far, predominantly featuring CaM-dependent protein kinases and phosphatases
[38]. The two groups are characterized by a number of CaM-binding modes, namely 1–14, 1–5–10 and 1–10–16. However, CaM has also been shown to form Ca
2+-free complexes. This includes, for example, the CaM-NaV1.5 (IQ-motif) interaction that occurs within the C-lobe of CaM and IQ-motif in α6 of NaV1.5 or the previously mentioned interactions with neurogranin and PITR. Notably, in these cases, the CaM C-lobe adopts a semi-open conformation
[39]. Another Ca
2+-free complex is CaM-Myosin-5A with the following topology: the C-lobes of the two CaM molecules bind to the N-terminals of the IQ motifs of NaV1.5 in an antiparallel orientation
[40]. The last type of CaM-targeted complexes features partial CaM saturation with Ca
2+. The CaM-binding domain (CaMBD) of small-conductance Ca
2+-activated potassium channel, SK2-a, forms a complex with a single CaM molecule at each of the two ends of the SK-CaMBD dimer consisting of two helix-loop-helix motifs. Interestingly, only the N-lobes of CaM molecules bound Ca
2+ wrapped around three α-helices of CaMBDs
[41]. The ApoCaM-SK2-a binding mode is different: a single SK CaMBD binds to the C-lobe of CaM, resulting in 1:1 stoichiometry. A helical fragment of the CaMBD was shown to interact with one of the CaM C-lobes
[42].
Here, our structural analysis showed that the interaction with FTO destabilized the CaM, especially the calcium-binding regions; nevertheless, the reciprocal effect was negligible. This may be explained by FTO homodimerization, when interfaces of the FTO–FTO homodimer and the FTO–CaM heterodimer are similar
[28]. Possibly, both interactions require FTO to maintain the same particular structure. Further, FTO–CaM interaction on the nanomolar level was also Ca
2+-dependent, with negligible
KD in the absence of Ca
2+. This feature is typical for CaM interaction with numerous partners
[43]. Importantly, this indicates that the proteins are likely to form a complex when the concentration of free Ca
2+ is sufficiently high and that they may be regulated by calcium concentration. For example, such complexes may be present in neural cells where the CaM level influences synapse development
[35] and FTO activity regulates neuronal development and the proliferation and differentiation of adult neural stem cells
[27].
Finally, we modelled the structure of the newly discovered FTO–CaM complex. The generated structure indicates similarity between the FTO–CaM-binding mode and that of the previously mentioned SK2-a-CaM. Indeed, FTO–CaM interaction at nanomolar concentrations is Ca2+-driven, but only two Ca2+ binding to the single lobe of CaM participate in the complex formation, while the second lobe remain free of Ca2+ (Figure 2a). This model is also consistent with the HDX results. The predicted stoichiometry of the biological FTO–CaM complex is 1:1, with the CaM molecule wrapped around the FTO C-terminal domain. Importantly, this type of interaction suggests that CaM–FTO may mediate FTO interaction with other partners.
The discovery of this exclusive CaM–FTO interaction type sheds a new light on the results of previous FTO studies
[8]. It is worth mentioning that both proteins interact with numerous other proteins, including three isoforms of CaMKII. Li and coworkers have shown that an increased FTO expression delays forskolin-mediated dephosphorylation of one of CaMKII’s cellular targets: cAMP response element-binding protein (CREB) in human neuroblastoma cells
[8]. However, direct influence of FTO on CaMKII enzymatic activity as well as FTO phosphorylation by this kinase were not observed. Due to the fact that CaM is a CaMKII activator, our results show new connections between CaM, CaMKII and FTO phosphorylation by CaMKII. Non-physiological FTO concentration of 25 µM in the performed experiment and the finding that the affinity of CaM/Ca
2+ to FTO seems to be higher than to CaMKII likely results in a shortage of free CaM necessary for CaMKII activation. Clearly, this relationship indicates that interplay between FTO, CaM and CaMKII needs further study
[8]. Finally, it cannot be excluded that FTO–CaM interaction may affect the binding of other FTO interactors. For example, as previously mentioned, the FTO C-terminal domain interacts with SFPQ protein. This interaction affects substrate specificity of the FTO protein, thus the regulatory role of calmodulin has to be taken into consideration.
Summing up, the last 10 years of intensive research into the role of the FTO protein in metabolism have identified a number of transcripts as substrates of this demethylase, affecting cell metabolism and mediating FTO specificity. Our discovery of CaM as another FTO interactor supports the thesis that this demethylase is regulated by other proteins. We discovered the ability of FTO, especially its C-terminal domain, to interact in vitro with the single lobe of a CaM molecule in a Ca2+-dependent manner. Moreover, only one part of CaM interacts with FTO, whereas the other one remains free. This interaction may be a part of regulatory mechanisms that FTO is subjected to, due to the fact that other FTO interactors, such as FTO itself and SFPQ protein, also bind to the FTO C-domain. The recognition of the role of CaM in the FTO interaction helps broaden the understanding of the function of this RNA modification demethylase. This finding is important because the multiplicity of FTO interactors may explain the involvement of this protein in a number of different metabolic pathways.
This entry is adapted from the peer-reviewed paper 10.3390/ijms221910869