MicroRNAs (miRNAs) are small non-coding RNAs that are about 22 nucleotides in length. They regulate gene expression post-transcriptionally with the effector protein complex, containing Argonaute or trinucleotide repeat containing 6 (TNRC6) proteins, and target mRNAs in a sequence-dependent manner, causing the translational repression and destabilization of the target mRNAs. Both Drosha and Dicer, members of the RNase III family proteins, are essential components in the canonical miRNA biogenesis pathway. miRNA is transcribed into primary-miRNA (pri-miRNA) from genomic DNA. Drosha then cleaves the flanking regions of pri-miRNA into precursor-miRNA (pre-miRNA), while Dicer cleaves the loop region of the pre-miRNA to form a miRNA duplex. In this report, we summarized and discussed the current reports in which double-stranded RNA binding proteins (dsRBPs), such as TAR RNA binding protein (TRBP) or the adenosine deaminase acting on RNA (ADAR), modulate the processing of miRNA by Dicer in various manners.
- microRNA biogenesis
- Dicer-associated proteins
- dsRBP
- TRBP
- ADAR
- PACT
- LGP2
- miRNA–mRNA network
1. Introduction
MicroRNAs (miRNAs) are single-stranded RNAs of approximately 22 nucleotides in length and are classified as small non-coding RNAs. The miRNAs regulate gene expression post-transcriptionally by a mechanism known as RNA silencing, where miRNA is loaded onto Argonaute (AGO), a core component of the miRNA-induced silencing complex (miRISC) [1]. While on AGO, the miRNA recognizes target mRNAs that have sequences that are complementary to the “seed region” (positions 2–8 from the 5′ end) of the miRNA in their 3′ untranslated region [2]. As the seed region only consists of seven nucleotides, each miRNA is capable of recognizing and regulating many types of mRNAs, indicating that miRNA–mRNA gene expression networks are highly complicated.
Some of the miRNAs that have been discovered in diverse eukaryotes are evolutionally conserved, while others are species specific [3][4][5][6]. The first two miRNAs, lin-4 and let-7, were discovered in Caenorhabditis elegans ( C. elegans ) through the analysis of the heterochronic gene mutants that undergo development/differentiation at an abnormal time within the organism [7][8][9]. Let-7 is evolutionally conserved across various species, including in humans. In human lung cancer cells, let-7 regulates cell proliferation and also suppresses the expression levels of NRAS and KRAS, two genes that induce oncogenic transformation when mutated [10][11][12]. Thus, although let-7 is conserved in both C. elegans and humans, its function in the two species is different.
At the initial stage of miRNA biogenesis, miRNA is transcribed by RNA polymerase II (Pol II) as primary-miRNA (pri-miRNA) , which has stem loop structures [13][14] (Figure 1). In the canonical miRNA biogenesis pathway, the flanking regions of the pri-miRNA are cleaved to generate precursor-miRNA (pre-miRNA) in the nucleus by a microprocessor complex consisting of Drosha, a member of the RNase III family proteins, and its cofactor DiGeorge syndrome critical region 8 (DGCR8), a double-stranded RNA (dsRNA) binding protein (dsRBP) [15][16][17]. The pre-miRNA is transported from the nucleus to the cytoplasm by Exportin-5 (EXP5), which then couples with GTP-bound Ran [18]. In the cytoplasm, Dicer, an RNase III family protein, cleaves off the loop region of the pre-miRNA to generate a miRNA duplex in collaboration with the trans-activation response (TAR) RNA binding protein (TRBP) in the canonical miRNA biogenesis pathway [19][20]. The interaction of the Dicer–TRBP complex with Argonaute (AGO) facilitates the loading of the miRNA duplex onto AGO to form the RISC-loading complex (RLC) [21][22][23]. The miRNA duplex is then unwound into single stranded miRNAs; the RNA strand that remains on the AGO protein acts as the miRNA, while the other strand is discarded [24]. The former is called the guide strand, and the latter is called the passenger strand. The mature miRNA on the AGO protein guides the RISC to target mRNAs that have sequences that are complementary to the seed region of the miRNA. After binding to the mRNA, AGO recruits the trinucleotide repeat containing 6 (TNRC6) protein, a scaffold protein tethering effector proteins to destabilize and translationally repress target mRNAs by inducing their decapping and deadenylation [25].
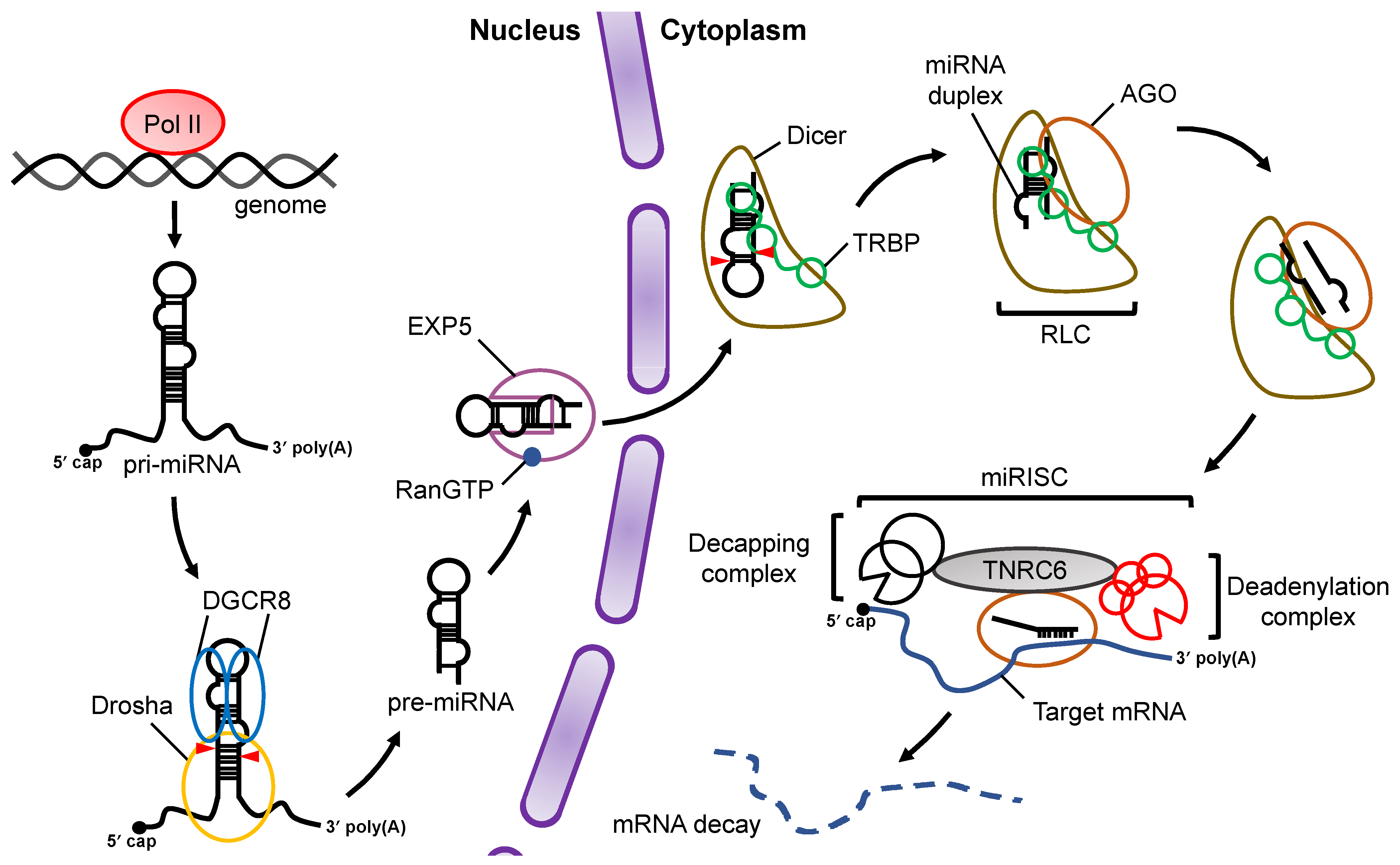 Figure 1. Overview of miRNA biogenesis and the RNA silencing pathway. Pri-miRNAs are transcribed from the genome by Pol II. In the nucleus, pri-miRNA is cleaved by a microprocessor complex consisting of Drosha and DGCR8 to produce pre-miRNA, which is then transported from the nucleus to the cytoplasm by EXP5 coupled with GTP-bound Ran (RanGTP). Cleavage of pre-miRNA is performed by Dicer and its cofactor, TRBP, in a canonical miRNA biogenesis pathway. After pre-miRNA cleavage, the miRNA duplex is loaded onto AGO proteins through formation of the RLC complex. The mature miRNA guides the RISC complex to target mRNAs that are complementary to the seed region of miRNA. Recruitment of the TNRC6 protein induces the destabilization and translational repression of the target mRNA.
Figure 1. Overview of miRNA biogenesis and the RNA silencing pathway. Pri-miRNAs are transcribed from the genome by Pol II. In the nucleus, pri-miRNA is cleaved by a microprocessor complex consisting of Drosha and DGCR8 to produce pre-miRNA, which is then transported from the nucleus to the cytoplasm by EXP5 coupled with GTP-bound Ran (RanGTP). Cleavage of pre-miRNA is performed by Dicer and its cofactor, TRBP, in a canonical miRNA biogenesis pathway. After pre-miRNA cleavage, the miRNA duplex is loaded onto AGO proteins through formation of the RLC complex. The mature miRNA guides the RISC complex to target mRNAs that are complementary to the seed region of miRNA. Recruitment of the TNRC6 protein induces the destabilization and translational repression of the target mRNA.
2. Processing of Pre-miRNA by Dicer
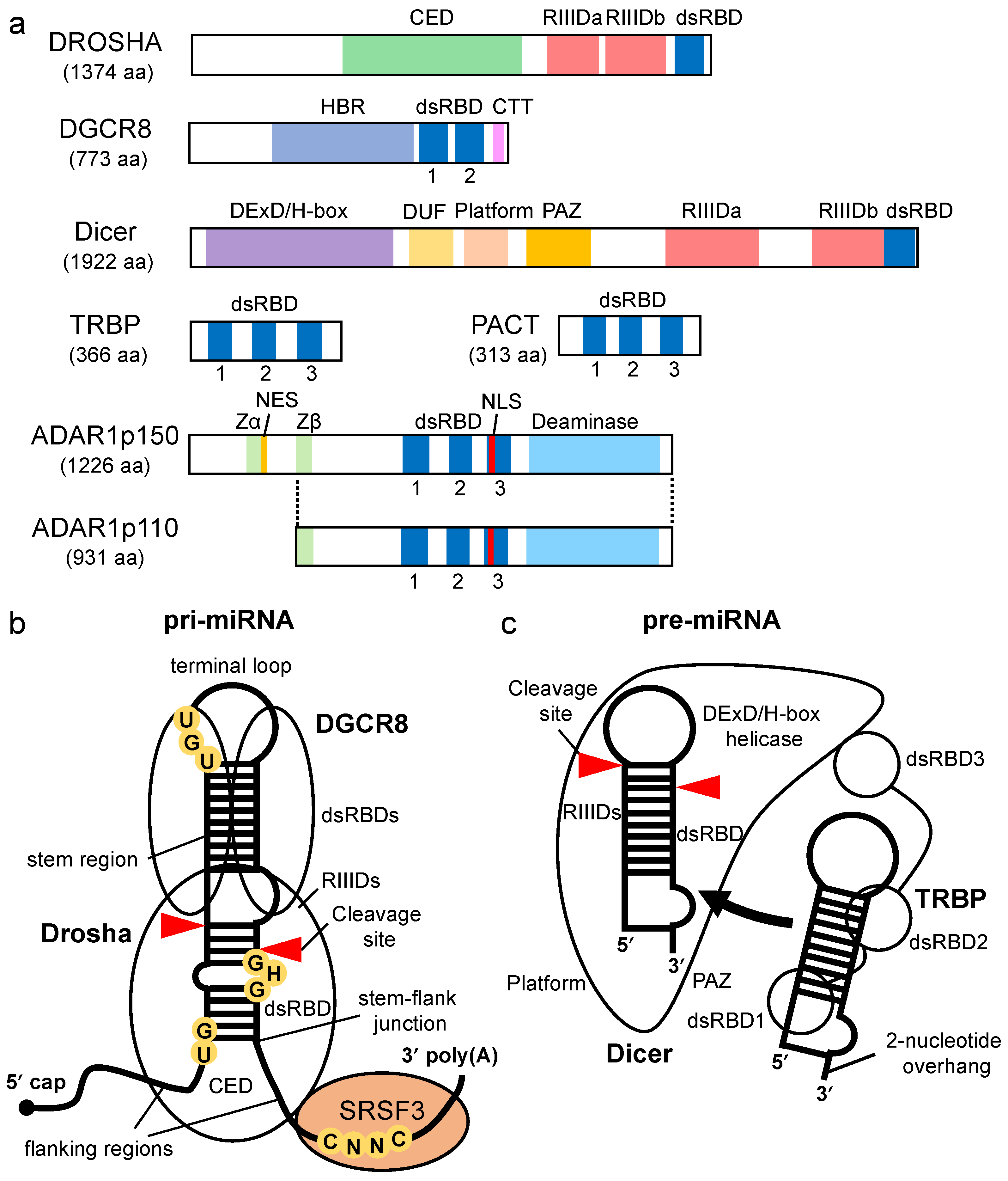
3. Enhancement of Pre-miRNA Processing by Dicer via TRBP/PACT
Dicer-associated proteins regulate the substrate recruitment and the cleavage activity of Dicer [43][44]. The processing of pre-miRNA is not only promoted by TRBP but also by other Dicer-associated proteins, such as PACT, a protein activator of protein kinase R (PKR) [43][45][46]. TRBP is a protein that binds to TAR RNA, a hairpin-structured RNA that is encoded by human immunodeficiency virus type I [47]. PACT is a protein that was initially identified as an activator of PKR [48]. TRBP and PACT have highly conserved domain structures with three dsRBDs [49] (Figure 2a). The dsRBDs are divided into two subclasses, type-A and type-B. Type-A has a conserved αβββα motif with a high affinity to dsRNA [50][51]. Type-B, also termed half dsRBD, has poorly conserved N-terminal sequences in the αβββα motif and is associated with protein–protein interactions [52][53]. The first and second dsRBDs of TRBP and PACT are type-As, and the third dsRBD is a type-B that binds to Dicer through the DExD/H-box helicase domain [43][45][46].
Both TRBP and PACT interact with Dicer to promote the cleavage of the pre-miRNA in the RLC containing AGO protein and the facilitate loading of the miRNA duplex onto AGO [20][23][43][54][55]. It was reported that deletion or mutation of the DExD/H-box helicase domain of Dicer activated the cleavage of its substrates, which suggested that this domain inhibits catalytic activity rather than affecting RNA-substrate binding [56]. TRBP binds to the DExD/H-box helicase domain of Dicer and stimulates the cleavage activity of Dicer. Therefore, the DExD/H-box helicase domain functions as an intramolecular structural switch that maintains Dicer in a low-activity state until the partner proteins interact with its DExD/H-box helicase domain. In addition, it was reported that TRBP facilitates the processing activity of pre-miRNA by Dicer in RNA-crowded molecular environments [57] and that it also facilitates the recruitment of pre-miRNAs to the PAZ domain of Dicer. Furthermore, the sliding motion of TRBP on dsRNA with Dicer has been reported [58]. This was associated with the higher substrate cleavage activity of Dicer compared to Dicer alone, which suggests that TRBP facilitates the cleavage activity of Dicer by guiding Dicer to the substrates. To date, no studies on the mechanism by which PACT promotes Dicer-mediated cleavage of pre-miRNAs have been reported. However, the amino acid sequence of the Dicer-interacting dsRBD of PACT was found to be similar to that of TRBP. It has yet to be determined if PACT interacts with Dicer by a mechanism similar to that of TRBP and if it enhances the processing of similar types of pre-miRNAs.
TRBP and PACT have different functions. Although TRBP preferentially binds to simple duplex RNA, PACT inhibits Dicer-mediated dsRNA cleavage for siRNA production [59]. Unlike PACT, the cleavage site for Dicer-TRBP shifts when compared to cleavage by Dicer alone [60][61]. PACT and TRBP have no redundant effects on the production of isomiRs , different-sized miRNAs that alter the downstream target-binding specificities. Such differences in dsRNA recognition and processing behavior are attributed to two N-terminal RNA-binding domains in each protein.
Several studies have addressed the TRBP-mediated maturation of specific miRNAs and its effect on downstream pathways. It was reported that the TRBP-mediated maturation of miR-208a decreased the expression level of SRY-Box Transcription Factor 6 (Sox6), which is required for normal heart function [62]. It was also reported that disruptions of TRBP-dependent maturations of tumor suppressor certain miRNAs (TS-miRs), miR-143 and miR-145, were related to the self-renewal and tumor maintenance of cancer stem cells [63]. These results suggest that TRBP regulates biogenesis and the downstream gene regulatory pathways of specific miRNAs.
4. Enhancement of Pre-miRNA Processing by Dicer via ADAR1
5. TRBP-LGP2 Interaction Inhibits Pre-miRNA Processing by Dicer
In virus infected mammalian cells, virus-derived RNAs are captured by viral sensor proteins such as retinoic acid-inducible gene I-like receptors (RLRs), inducing the production of type I interferon [74][75][76]. Recently, we reported that the TRBP-mediated maturations of pre-miRNAs were inhibited through the competitive binding of the laboratory of genetics and physiology 2 (LGP2) to Dicer–TRBP interaction during Sendai virus infection [77]. Interferons enhanced the expression of LGP2, which interacted with TRBP to inhibit the Dicer–TRBP interaction [78]. Following LGP2-dependent inhibition of Dicer–TRBP interaction, the maturations of TRBP-bound pre-miRNAs, including miR-106b, were suppressed. The inhibition of the maturation of such miRNAs increased the expression of apoptosis-related genes downstream of miRNA processing. This finding suggested that the crosstalk between antiviral response and miRNA biogenesis is regulated by TRBP binding to the specific pre-miRNAs.
This entry is adapted from the peer-reviewed paper 10.3390/ncrna7030057
References
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296.
- Grimson, A.; Farh, K.K.-H.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105.
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89.
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858.
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862.
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864.
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862.
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854.
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906.
- Johnson, C.D.; Esquela-Kerscher, A.; Stefani, G.; Byrom, M.; Kelnar, K.; Ovcharenko, D.; Wilson, M.; Wang, X.; Shelton, J.; Shingara, J.; et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007, 67, 7713–7722.
- Takamizawa, J.; Konishi, H.; Yanagisawa, K.; Tomida, S.; Osada, H.; Endoh, H.; Harano, T.; Yatabe, Y.; Nagino, M.; Nimura, Y.; et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004, 64, 3753–3756.
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647.
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966.
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060.
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419.
- Denli, A.M.; Tops, B.B.J.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235.
- Gregory, R.I.; Yan, K.-P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240.
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98.
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838.
- Haase, A.D.; Jaskiewicz, L.; Zhang, H.; Lainé, S.; Sack, R.; Gatignol, A.; Filipowicz, W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005, 6, 961–967.
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640.
- Maniataki, E.; Mourelatos, Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005, 19, 2979–2990.
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744.
- Kawamata, T.; Seitz, H.; Tomari, Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 2009, 16, 953–960.
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433.
- Lee, Y.S.; Nakahara, K.; Pham, J.W.; Kim, K.; He, Z.; Sontheimer, E.J.; Carthew, R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 2004, 117, 69–81.
- Fukudome, A.; Fukuhara, T. Plant dicer-like proteins: Double-stranded RNA-cleaving enzymes for small RNA biogenesis. J. Plant Res. 2017, 130, 33–44.
- Vermeulen, A.; Behlen, L.; Reynolds, A.; Wolfson, A.; Marshall, W.S.; Karpilow, J.; Khvorova, A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA 2005, 11, 674–682.
- Zhang, H.; Kolb, F.A.; Brondani, V.; Billy, E.; Filipowicz, W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002, 21, 5875–5885.
- Taylor, D.W.; Ma, E.; Shigematsu, H.; Cianfrocco, M.A.; Noland, C.L.; Nagayama, K.; Nogales, E.; Doudna, J.A.; Wang, H.-W. Substrate-specific structural rearrangements of human Dicer. Nat. Struct. Mol. Biol. 2013, 20, 662–670.
- Gu, S.; Jin, L.; Zhang, Y.; Huang, Y.; Zhang, F.; Valdmanis, P.N.; Kay, M.A. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell 2012, 151, 900–911.
- Lau, P.-W.; Potter, C.S.; Carragher, B.; MacRae, I.J. Structure of the human Dicer-TRBP complex by electron microscopy. Structure 2009, 17, 1326–1332.
- Wang, H.-W.; Noland, C.; Siridechadilok, B.; Taylor, D.W.; Ma, E.; Felderer, K.; Doudna, J.A.; Nogales, E. Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 2009, 16, 1148–1153.
- Macrae, I.J.; Zhou, K.; Li, F.; Repic, A.; Brooks, A.N.; Cande, W.Z.; Adams, P.D.; Doudna, J.A. Structural basis for double-stranded RNA processing by Dicer. Science 2006, 311, 195–198.
- MacRae, I.J.; Zhou, K.; Doudna, J.A. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007, 14, 934–940.
- Park, J.-E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205.
- Tian, Y.; Simanshu, D.K.; Ma, J.-B.; Park, J.-E.; Heo, I.; Kim, V.N.; Patel, D.J. A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol. Cell 2014, 53, 606–616.
- Blaszczyk, J.; Tropea, J.E.; Bubunenko, M.; Routzahn, K.M.; Waugh, D.S.; Court, D.L.; Ji, X. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure 2001, 9, 1225–1236.
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68.
- Tsutsumi, A.; Kawamata, T.; Izumi, N.; Seitz, H.; Tomari, Y. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat. Struct. Mol. Biol. 2011, 18, 1153–1158.
- Sinha, N.K.; Iwasa, J.; Shen, P.S.; Bass, B.L. Dicer uses distinct modules for recognizing dsRNA termini. Science 2018, 359, 329–334.
- Wostenberg, C.; Lary, J.W.; Sahu, D.; Acevedo, R.; Quarles, K.A.; Cole, J.L.; Showalter, S.A. The role of human Dicer-dsRBD in processing small regulatory RNAs. PLoS ONE 2012, 7, e51829.
- Lee, Y.; Hur, I.; Park, S.-Y.; Kim, Y.-K.; Suh, M.R.; Kim, V.N. The role of PACT in the RNA silencing pathway. EMBO J. 2006, 25, 522–532.
- Kim, Y.; Yeo, J.; Lee, J.H.; Cho, J.; Seo, D.; Kim, J.-S.; Kim, V.N. Deletion of human tarbp2 reveals cellular microRNA targets and cell-cycle function of TRBP. Cell Rep. 2014, 9, 1061–1074.
- Daniels, S.M.; Melendez-Peña, C.E.; Scarborough, R.J.; Daher, A.; Christensen, H.S.; El Far, M.; Purcell, D.F.J.; Lainé, S.; Gatignol, A. Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol. Biol. 2009, 10, 38.
- Wilson, R.C.; Tambe, A.; Kidwell, M.A.; Noland, C.L.; Schneider, C.P.; Doudna, J.A. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol. Cell 2015, 57, 397–407.
- Gatignol, A.; Buckler-White, A.; Berkhout, B.; Jeang, K.T. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science 1991, 251, 1597–1600.
- Patel, R.C.; Sen, G.C. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998, 17, 4379–4390.
- Saito, K.; Ishizuka, A.; Siomi, H.; Siomi, M.C. Processing of pre-microRNAs by the Dicer-1–Loquacious COMPLEX in Drosophila cells. PLoS Biol. 2005, 3, e235.
- Bycroft, M.; Grünert, S.; Murzin, A.G.; Proctor, M.; St Johnston, D. NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J. 1995, 14, 3563–3571.
- Kharrat, A.; Macias, M.J.; Gibson, T.J.; Nilges, M.; Pastore, A. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 1995, 14, 3572–3584.
- Krovat, B.C.; Jantsch, M.F. Comparative mutational analysis of the double-stranded RNA binding domains of Xenopus laevis RNA-binding protein A. J. Biol. Chem. 1996, 271, 28112–28119.
- Hitti, E.G.; Sallacz, N.B.; Schoft, V.K.; Jantsch, M.F. Oligomerization activity of a double-stranded RNA-binding domain. FEBS Lett. 2004, 574, 25–30.
- Ota, H.; Sakurai, M.; Gupta, R.; Valente, L.; Wulff, B.-E.; Ariyoshi, K.; Iizasa, H.; Davuluri, R.V.; Nishikura, K. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 2013, 153, 575–589.
- Chakravarthy, S.; Sternberg, S.H.; Kellenberger, C.A.; Doudna, J.A. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J. Mol. Biol. 2010, 404, 392–402.
- Ma, E.; MacRae, I.J.; Kirsch, J.F.; Doudna, J.A. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 2008, 380, 237–243.
- Fareh, M.; Yeom, K.-H.; Haagsma, A.C.; Chauhan, S.; Heo, I.; Joo, C. TRBP ensures efficient Dicer processing of precursor microRNA in RNA-crowded environments. Nat. Commun. 2016, 7, 13694.
- Koh, H.R.; Kidwell, M.A.; Ragunathan, K.; Doudna, J.A.; Myong, S. ATP-independent diffusion of double-stranded RNA binding proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 151–156.
- Lee, H.Y.; Zhou, K.; Smith, A.M.; Noland, C.L.; Doudna, J.A. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013, 41, 6568–6576.
- Lee, H.Y.; Doudna, J.A. TRBP alters human precursor microRNA processing in vitro. RNA 2012, 18, 2012–2019.
- Fukunaga, R.; Han, B.W.; Hung, J.-H.; Xu, J.; Weng, Z.; Zamore, P.D. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell 2012, 151, 533–546.
- Ding, J.; Chen, J.; Wang, Y.; Kataoka, M.; Ma, L.; Zhou, P.; Hu, X.; Lin, Z.; Nie, M.; Deng, Z.-L.; et al. Trbp regulates heart function through microRNA-mediated Sox6 repression. Nat. Genet. 2015, 47, 776–783.
- De Vito, C.; Riggi, N.; Cornaz, S.; Suvà, M.-L.; Baumer, K.; Provero, P.; Stamenkovic, I. A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer Cell 2012, 21, 807–821.
- Bass, B.L.; Weintraub, H. A developmentally regulated activity that unwinds RNA duplexes. Cell 1987, 48, 607–613.
- Bass, B.L.; Weintraub, H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 1988, 55, 1089–1098.
- Wagner, R.W.; Smith, J.E.; Cooperman, B.S.; Nishikura, K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. USA 1989, 86, 2647–2651.
- Yang, W.; Chendrimada, T.P.; Wang, Q.; Higuchi, M.; Seeburg, P.H.; Shiekhattar, R.; Nishikura, K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006, 13, 13–21.
- Kawahara, Y.; Zinshteyn, B.; Chendrimada, T.P.; Shiekhattar, R.; Nishikura, K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer–TRBP complex. EMBO Rep. 2007, 8, 763–769.
- Reich, D.P.; Tyc, K.M.; Bass, B.L. C. elegans ADARs antagonize silencing of cellular dsRNAs by the antiviral RNAi pathway. Genes Dev. 2018, 32, 271–282.
- Montavon, T.C.; Lefèvre, M.; Baldaccini, M.; Girardi, E.; Chane-Woon-Ming, B.; Messmer, M.; Hammann, P.; Chicher, J.; Pfeffer, S. Human Dicer helicase domain acts as an interaction platform to recruit PKR and dsRNA binding proteins during viral infection. Cold Spring Harb. Lab. 2020, 2020.
- Wang, Q.; Miyakoda, M.; Yang, W.; Khillan, J.; Stachura, D.L.; Weiss, M.J.; Nishikura, K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 2004, 279, 4952–4961.
- Zhang, X.; Gao, X.; Hu, J.; Xie, Y.; Zuo, Y.; Xu, H.; Zhu, S. ADAR1p150 Forms a Complex with Dicer to Promote miRNA-222 Activity and Regulate PTEN Expression in CVB3-Induced Viral Myocarditis. Int. J. Mol. Sci. 2019, 20, 407.
- Liu, X.; Fu, Y.; Huang, J.; Wu, M.; Zhang, Z.; Xu, R.; Zhang, P.; Zhao, S.; Liu, L.; Jiang, H. ADAR1 promotes the epithelial-to-mesenchymal transition and stem-like cell phenotype of oral cancer by facilitating oncogenic microRNA maturation. J. Exp. Clin. Cancer Res. 2019, 38, 315.
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 2001, 413, 732–738.
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737.
- Yoneyama, M.; Kikuchi, M.; Matsumoto, K.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Foy, E.; Loo, Y.-M.; Gale, M., Jr.; Akira, S.; et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005, 175, 2851–2858.
- Takahashi, T.; Nakano, Y.; Onomoto, K.; Yoneyama, M.; Ui-Tei, K. LGP2 virus sensor enhances apoptosis by upregulating apoptosis regulatory genes through TRBP-bound miRNAs during viral infection. Nucleic Acids Res. 2020, 48, 1494–1507.
- Takahashi, T.; Nakano, Y.; Onomoto, K.; Murakami, F.; Komori, C.; Suzuki, Y.; Yoneyama, M.; Ui-Tei, K. LGP2 virus sensor regulates gene expression network mediated by TRBP-bound microRNAs. Nucleic Acids Res. 2018, 46, 9134–9147.
