Enterococci derived from an ancestor that was a commensal of aquatic life forms, when animals first became terrestrial. To cope with the stresses of the new terrestrial habitat, enterococci evolved to be tough bugs, resistant to a wide range of environmental and host factors. That made them extremely successful not only in adapting to the new way of life of their hosts, but also in colonizing other non-animal, and even inanimate, environments - such as feeds and foods. The plasticity of the enterococcal genome, together with their notable ability to trade virulence and antibiotic resistance, have enabled them to also become notable opportunistic, multi-resistant pathogens and act as reservoirs of pathogenicity and resistance determinants.
- Enterococcus
- enterococci
- commensal
- opportunistic pathogen
- antibiotic resistance
1. Introduction
2. Persistence of VRE Strains in Portuguese Food Animals, an Example
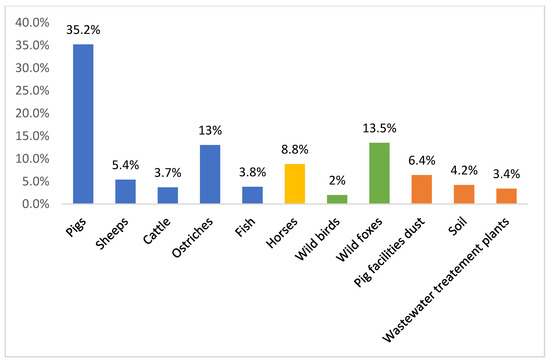
3. Findings
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms8081118
References
- Wendy Escobedo-Hinojosa; Liliana Pardo-López; Analysis of bacterial metagenomes from the Southwestern Gulf of Mexico for pathogens detection. Pathogens and Disease 2017, 75, 103430, 10.1093/femspd/ftx058.
- Wendy Escobedo-Hinojosa; Liliana Pardo-López; Analysis of bacterial metagenomes from the Southwestern Gulf of Mexico for pathogens detection. Pathogens and Disease 2017, 75, ftx058, 10.1093/femspd/ftx058.
- Carolina Baldisserotto Comerlato; Ana Carolina Ritter; Kendi Nishino Miyamoto; Adriano Brandelli; Proteomic study of Enterococcus durans LAB18S growing on prebiotic oligosaccharides. Food Microbiology 2020, 89, 103430, 10.1016/j.fm.2020.103430.
- O. Nilsson; Vancomycin resistant enterococci in farm animals – occurrence and importance. Infection Ecology & Epidemiology 2012, 2, 606, 10.3402/iee.v2i0.16959.
- Guido Werner; Teresa M. Coque; Charles M.A.P. Franz; Elisabeth Grohmann; Kristin Hegstad; L.B. Jensen; Willem Van Schaik; Keith Weaver; Antibiotic resistant enterococci—Tales of a drug resistance gene trafficker. International Journal of Medical Microbiology 2013, 303, 360-379, 10.1016/j.ijmm.2013.03.001.
- Krista Dubin; Eric G. Pamer; Enterococci and Their Interactions with the Intestinal Microbiome. Mobile DNA III 2018, 5, 309-330, 10.1128/microbiolspec.bad-0014-2016.
- Rahat Zaheer; Shaun R. Cook; Ruth Barbieri; Noriko Goji; Andrew Cameron; Aaron Petkau; Rodrigo Ortega Polo; Lisa Tymensen; Courtney Stamm; Jiming Song; et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Scientific Reports 2020, 10, 1-16, 10.1038/s41598-020-61002-5.
- Carmen Torres; Carla Alonso; Laura Ruiz-Ripa; Ricardo Leon-Sampedro; Rosa Del Campo; Teresa M. Coque; Antimicrobial Resistance in Enterococcus spp. of animal origin. Antimicrobial Resistance in Bacteria from Livestock and Companion Animals 2018, 6, 185-227, 10.1128/microbiolspec.arba-0032-2018.
- Xiaoxiao Qiao; Renpeng Du; Yu Wang; Ye Han; Zhijiang Zhou; Isolation, Characterisation and Fermentation Optimisation of Bacteriocin-Producing Enterococcus faecium. Waste and Biomass Valorization 2019, 11, 1-9, 10.1007/s12649-019-00634-9.
- K. Fisher; Carol Phillips; The ecology, epidemiology and virulence of Enterococcus. Microbiology 2009, 155, 1749-1757, 10.1099/mic.0.026385-0.
- Carolina Vieira De Almeida; Antonio Taddei; Amedeo Amedei; The controversial role of Enterococcus faecalis in colorectal cancer. Therapeutic Advances in Gastroenterology 2018, 11, 1756284818783606, 10.1177/1756284818783606.
- Talat A. El-Kersh; Mohammed A. Marie; Yazeed A. Al-Sheikh; Mohamed H.M. Al-Agamy; Ahmad A. Al Bloushy; Prevalence and risk factors of early fecal carriage of Enterococcus faecalis and Staphylococcus spp and their antimicrobial resistant patterns among healthy neonates born in a hospital setting in central Saudi Arabia. Saudi Medical Journal 2016, 37, 280-287, 10.15537/smj.2016.3.13871.
- Liu, L.; Wang, Q.; Wu, X.; Qi, H.; Das, R.; Lin, H.; Shi, J.; Wang, S.; Yang, J.; Xue, Y.; et al. Vancomycin exposure caused opportunistic pathogens bloom in intestinal microbiome by simulator of the human intestinal microbial ecosystem (SHIME). Environmental Pollution 2020, 265, 114399, j.envpol.2020.114399.
- Cesar A. Arias; Barbara E. Murray; The rise of the Enterococcus: beyond vancomycin resistance. Nature Reviews Genetics 2012, 10, 266-278, 10.1038/nrmicro2761.
- Antoni P.A. Hendrickx; Willem Van Schaik; Rob J.L. Willems; The cell wall architecture ofEnterococcus faecium: from resistance to pathogenesis. Future Microbiology 2013, 8, 993-1010, 10.2217/fmb.13.66.
- Mónica García-Solache; Louis B. Rice; The Enterococcus: a Model of Adaptability to Its Environment. Clinical Microbiology Reviews 2019, 32, e00058-18, 10.1128/cmr.00058-18.
- Mutshiene Deogratias Ekwanzala; John Barr Dewar; Ilunga Kamika; Maggy Ndombo Benteke Momba; Comparative genomics of vancomycin-resistant Enterococcus spp. revealed common resistome determinants from hospital wastewater to aquatic environments. Science of The Total Environment 2020, 719, 137275, 10.1016/j.scitotenv.2020.137275.
- Sónia Ramos; Gilberto Igrejas; J. Rodrigues; José L. Capelo-Martínez; Patrícia Poeta; Genetic characterisation of antibiotic resistance and virulence factors in vanA-containing enterococci from cattle, sheep and pigs subsequent to the discontinuation of the use of avoparcin. The Veterinary Journal 2012, 193, 301-303, 10.1016/j.tvjl.2011.12.007.
- Carlos Araújo; Carmen Torres; Alexandre Gonçalves; Catarina Carneiro; María López; Hajer Radhouani; Miguel Ângelo Pardal; Gilberto Igrejas; Patrícia Poeta; Genetic Detection and Multilocus Sequence Typing ofvanA-ContainingEnterococcusStrains from Mullets Fish (Liza ramada). Microbial Drug Resistance 2011, 17, 357-361, 10.1089/mdr.2010.0171.
- Carlos Araújo; Carmen Torres; Nuno Silva; Catarina Carneiro; Alexandre Gonçalves; Hajer Radhouani; Susana Correia; Paulo Martins Da Costa; Rui Pacheco; Myriam Zarazaga; et al. Vancomycin-resistant enterococci from Portuguese wastewater treatment plants. Journal of Basic Microbiology 2010, 50, 605-609, 10.1002/jobm.201000102.
- Alexandre Gonçalves; Patrícia Poeta; Nuno Silva; Carlos Araújo; María López; Elena Ruiz; Inna Uliyakina; João Direitinho; Gilberto Igrejas; Carmen Torres; et al. Characterization of Vancomycin-Resistant Enterococci Isolated from Fecal Samples of Ostriches by Molecular Methods. Foodborne Pathogens and Disease 2010, 7, 1133-1136, 10.1089/fpd.2010.0548.
- Ines Moura; Hajer Radhouani; Carmen Torres; Patrícia Poeta; G. Igrejas; Detection and genetic characterisation of vanA-containing Enterococcus strains in healthy Lusitano horses. Equine Veterinary Journal 2010, 42, 181-183, 10.2746/042516409x480386.
- Hajer Radhouani; Gilberto Igrejas; Carlos Carvalho; Luís Pinto; Alexandre Gonçalves; María López; Roberto Sargo; Luís Cardoso; António Martinho; Vítor Rego; et al. Clonal Lineages, Antibiotic Resistance and Virulence Factors in Vancomycin-Resistant Enterococci Isolated from Fecal Samples of Red Foxes (Vulpes Vulpes). Journal of Wildlife Diseases 2011, 47, 769-773, 10.7589/0090-3558-47.3.769.
- Teresa M. Braga; Constança Pomba; Maria De Fátima Silva Lopes; High-level vancomycin resistant Enterococcus faecium related to humans and pigs found in dust from pig breeding facilities. Veterinary Microbiology 2012, 161, 344-349, 10.1016/j.vetmic.2012.07.034.
- Vanessa Silva; Gilberto Igrejas; Isabel Carvalho; Fernando Peixoto; Luís Cardoso; Jose E. Pereira; Rosa Del Campo; Patrícia Poeta; Genetic Characterization of vanA-Enterococcus faecium Isolates from Wild Red-Legged Partridges in Portugal. Microbial Drug Resistance 2017, 24, 89-94, 10.1089/mdr.2017.0040.
- Vanessa Silva; Fernando Peixoto; Gilberto Igrejas; Carolina Parelho; Patricia V. Garcia; Isabel Carvalho; Margarida Sousa; José Pereira; Armindo Dos Santos Rodrigues; Patrícia Poeta; et al. First Report on vanA-Enterococcus faecalis Recovered from Soils Subjected to Long-Term Livestock Agricultural Practices in Azores Archipelago. International Journal of Environmental Research 2018, 12, 39-44, 10.1007/s41742-018-0068-0.
- Frank M. Aarestrup; Characterization of Glycopeptide-Resistant Enterococcus faecium (GRE) from Broilers and Pigs in Denmark: Genetic Evidence that Persistence of GRE in Pig Herds Is Associated with Coselection by Resistance to Macrolides. Journal of Clinical Microbiology 1999, 38, 2774-2777, 10.1128/jcm.38.7.2774-2777.2000.
- Ana R. Freitas; Teresa M. Coque; Carla Novais; Anette M. Hammerum; Camilla H. Lester; Marcus J. Zervos; Susan Donabedian; L.B. Jensen; Maria Victoria Francia; Fernando Baquero; et al. Human and Swine Hosts Share Vancomycin-Resistant Enterococcus faecium CC17 and CC5 and Enterococcus faecalis CC2 Clonal Clusters Harboring Tn1546 on Indistinguishable Plasmids. Journal of Clinical Microbiology 2011, 49, 925-931, 10.1128/jcm.01750-10.
- Carolina Sabença; Telma De Sousa; Soraia Oliveira; Didier Viala; Laetitia Théron; Christophe Chambon; Michel Hébraud; Racha Beyrouthy; Richard Bonnet; Manuela Caniça; et al. Next-Generation Sequencing and MALDI Mass Spectrometry in the Study of Multiresistant Processed Meat Vancomycin-Resistant Enterococci (VRE). Biology 2020, 9, 89, 10.3390/biology9050089.
- Ramin Mazaheri Nezhad Fard; Payam Haghighi Khoshkhoo; Maryam Abbaspour; Zahra Rajabi; Study of VanA, B, C, D, E Genes in Vancomycin Resistant Enterococci Isolated from Retailed Dried Vegetables in Tehran, Iran. International Journal of Enteric Pathogens 2019, 7, 9-14, 10.15171/ijep.2019.03.
- Ali Jahansepas; Yaeghob Sharifi; Mohammad Aghazadeh; Mohammad Ahangarzadeh Rezaee; Comparative analysis of Enterococcus faecalis and Enterococcus faecium strains isolated from clinical samples and traditional cheese types in the Northwest of Iran: antimicrobial susceptibility and virulence traits. Archives of Microbiology 2019, 202, 765-772, 10.1007/s00203-019-01792-z.
- Ali Hassan Ahmed Al-Shammary; Run-off Patterns of Vancomycin Resistant Enterococci (VRE clones) in Cows Raw Milk and Imported Milk Powders at Baghdad Markets. Iraqi Journal of Veterinary Medicine 2019, 43, 61-66, 10.30539/iraqijvm.v43i2.532.
- Vanessa Pereira Perez Alonso; Murilo M Queiroz; Miriã L Gualberto; M.S. Nascimento; Klebsiella pneumonia carbapenemase (KPC), methicillin-resistant Staphylococcus aureus (MRSA), and vancomycin-resistant Enterococcus spp. (VRE) in the food production chain and biofilm formation on abiotic surfaces. Current Opinion in Food Science 2019, 26, 79-86, 10.1016/j.cofs.2019.04.002.
- Carla Novais; T. M. Coque; M. J. Costa; J. C. Sousa; F. Baquero; Luísa Peixe; High occurrence and persistence of antibiotic-resistant enterococci in poultry food samples in Portugal. Journal of Antimicrobial Chemotherapy 2005, 56, 1139-1143, 10.1093/jac/dki360.
- Ana R. Freitas; Carla Novais; Rosa Correia; Márcia Monteiro; Teresa M. Coque; Luísa Peixe; Non-susceptibility to tigecycline in enterococci from hospitalised patients, food products and community sources. International Journal of Antimicrobial Agents 2011, 38, 174-176, 10.1016/j.ijantimicag.2011.04.014.
- Joana Barbosa; Vânia Borges Ferreira; Paula Teixeira; Antibiotic susceptibility of enterococci isolated from traditional fermented meat products. Food Microbiology 2009, 26, 527-532, 10.1016/j.fm.2009.03.005.
- Ribeiro, T.; Oliveira, M.; Fraqueza, M.J.; Lauková, A.; Elias, M.; Tenreiro, R.; Barreto, A.S.; SemedoLemsaddek, R.; Antibiotic resistance and virulence factors among enterococci isolated from chouriço, a raditional Portuguese dry fermented sausage. Journal of Food Protection 2011, 74, 465–469, 10.4315/0362-028X.JFP- 10-309.
- M.F.P. Domingos-Lopes; Catherine Stanton; P.R. Ross; Maria De Lurdes Enes Dapkevicius; C.C. Silva; R. Paul Ross; Genetic diversity, safety and technological characterization of lactic acid bacteria isolated from artisanal Pico cheese. Food Microbiology 2017, 63, 178-190, 10.1016/j.fm.2016.11.014.
- Câmara, S.P.A.; Dapkevicius, A.; Silva, C.C.G.; Malcata, F.X.; Dapkevicius, M.L.N.E.; Artisanal Pico cheese as reservoir of Enterococcus species possessing virulence and antibiotic resistance properties: implications for food safety. Food Biotechnology 2020, 34, 25–41, 10.1080/08905436.2019.1710844.pira.
