Endophytic bacterial communities are beneficial communities for host plants that exist inside the surfaces of plant tissues, and their application improves plant growth. They benefit directly from the host plant by enhancing the nutrient amount of the plant’s intake and influencing the phytohormones, which are responsible for growth promotion and stress. Endophytic bacteria play an important role in plant-growth promotion (PGP) by regulating the indirect mechanism targeting pest and pathogens through hydrolytic enzymes, antibiotics, biocontrol potential, and nutrient restriction for pathogens.
1. Introduction
Plants can build a relationship with their ecosystem members to enhance growth and development in natural environments, as well as maintaining an ecological niche for thriving microbes. Numerous kinds of microorganisms, including bacterial and fungal communities, are harboring plant compartments that have been isolated and identified by using plant tissues and are nominated as endophytes. A majority of endophytes colonize with different parts of the plant apoplast, proliferating among the cellular spaces, including xylem vessels. Endophytes can also colonize plant reproductive organs, such as flower buds, flower petals, fruits, and seeds [
1]. For the first time, the isolation of the endophytic bacteria was carried out by Samish and Mund [
2] after surface sterilization of the plant tissues. Afterward, many researchers successfully categorized and reported more than 200 genera of endophytic bacteria from different plant tissues. These genera belong to the 16 phyla that are culturable and unculturable endophytes: Acidobacteria, Aquificae, Actinobacteria, Bacteroidetes, Chloroflexi, Chlorobi, Cyanobacteria, Proteobacteria, Firmicutes, Deinococcus-thermus, Fusobacteria, Planctomycetes, Gemmatimonates, Nitrospira, Spirochaetes, and Verrucomicrobia [
1,
3]. However, there is no such compressive report on phylum numbers identified from rice plant tissues. The previous literature revealed that several endophytes represent the three main phyla, namely Proteobacteria, Firmicutes, and Actinobacteria, comprising members of
Azoarcus [
4],
Bacillus [
5],
Enterobacter [
6],
Gluconobacter,
Stentrophomonas [
7],
Herbaspirillum [
8],
Pseudomonas,
Serratia [
9], and
Streptomyces [
10].
Rice (
Oryza sativa L.) is the most widely important grown cereal crop and is consumed by more than 50% of the world’s population [
11]. The ever-increasing world population demands sustainable agriculture production to feed 7.3 billion people; that population may become 9.7 billion up to 2064 (estimated by the United Nations (UN) [
12,
13]). An increasing population needs a high yield of rice to compete for this competition, but it must be achieved without, or by minimizing, the application of synthetic products due to high concern about environmental protection. Still, the main source of crop improvement in the agriculture sector is the practice of applying synthetic products, such as commercial fertilizers, nutrient supplements, insecticides, and pesticides. This action has hazardous effects on the environment and human and livestock health. The farmer communities are convinced to change their old practices by using alternative products that could be environmentally friendly [
14]. The bio-fertilizers and pesticides consist of non-pathogenic microorganisms; this uniqueness in nature facilitates attaining sustainable agriculture and environmental protection from chemical hazards. Additionally, bio-formulations might be the best alternative option to reduce environmental degradation and the threat to human health.
In the European Union Pesticides Database [
15], several registered bio-fertilizers and pesticides containing bacterial products are mainly synthesized by using
Bacillus species and
Pseudomonas species. In China, Fang [
16] reported the sources of registered bio-fertilizer products; these can be the single and combination of the bacterial strains of
Bacillus species (
B. licheniformis,
B. amyloliguefaciens,
B. megaterium, and
B. subtilis) and
Lactobacillus plantarum,
Paemibacillus mucilaginosus, and other species. These abovementioned members of genera are dominated among plants, soil, and under other environments, due to salient features, such as rapid proliferation and simple diet requirements, which can assist in easy propagation of bacteria in the environment. The above-highlighted bacteria are famous as beneficial organisms for plant growth, as well as for producing many metabolites to improve plant health. Therefore, these genera could be the best choice for preparing potential inoculants by mixing bacterial strains due to their diversified adaption to multiple environmental factors, such as pH, low and high temperature, salinity, and high metals, as well as the ability of their cells to remain alive in diverse environments for a long time [
17]. Importantly, the endophytic bacterial strains that are beneficial for growth promotion and disease suppression will be a better choice for making bio-preparations and minimize the risk of drawbacks and promote environmental protection. Endophytic bacteria can invade and colonize within plants in such a way that they are specific and remain intact. Moreover, strains of endophytic bacteria can enhance plant protection from phytopathogens and improve plant immunity.
The prominent abilities of endophytic bacteria to improve plant growth and minimize pathogen infestation via direct and indirect mechanisms [
18,
19] are attracting scientists globally. Direct mechanisms are set up to apply to those bacterial traits that directly promote plant growth. They include the production of Indole-acetic-acid (IAA), 1-aminocyclopane-1 carboxylic acid (ACC), and Gibberellic acids (GA), and also facilitate the uptake of fixed nitrogen, siderophore, phosphorus, and zinc production. Under indirect mechanisms, endophytic bacteria improve plant growth by acting as biocontrol agents. In addition, many endophytic bacteria have been reported to produce compounds classified as secondary metabolites for inhibition of pathogens, and they also adopt different mechanisms in order to produce cell wall degrading enzymes, antibiotics, competition and induced systemic resistance (ISR) [
20].
2. Endophytic Bacterial Diversity in the Rice Plant
To study the endophytic bacteria taxonomic flora in the rice plant, we collected the prehistoric taxonomic data until 2020, with their plant-growth-promoting traits and their antagonism against phytopathogens, using the National Centre of biotechnology information (NCBI) PubMed (
https://pubmed.ncbi.nlm.nih.gov/, accessed on 25 September 2020), Google Scholar (
https://scholar.google.com/, accessed on 25 September 2020), and SciencDirect (
https://www.sciencedirect.com/, accessed on 25 September 2020) web portals; after that, we plotted the taxonomic tree by writing a self-script in Python 2.7 [
21] to convert the excel file into Newick format; the further Newick-format file was visualized and edited in iTOL version 5 [
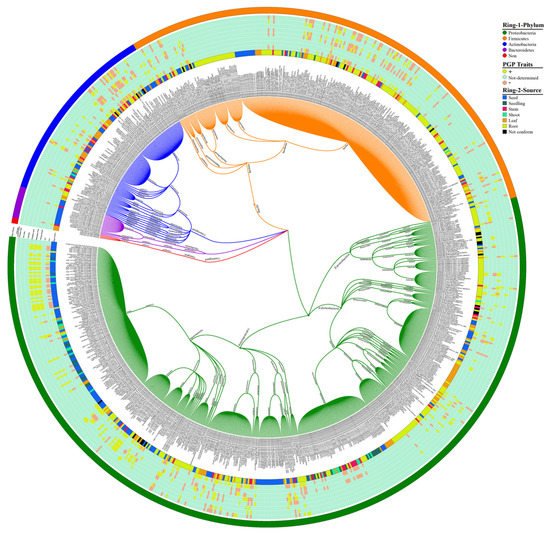
22]. Our data analysis showed 4 phyla, 6 classes, 22 orders, 46 families, and 74 genera of endophytic bacterial communities that have been mined from rice parts based on a culture-dependent method; complete details of taxonomic distribution are visualized in
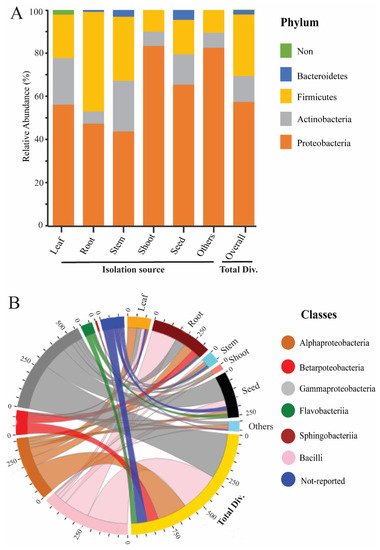
Figure 1. Moreover, our data analysis showed that the phylum Proteobacteria has a high abundance among rice tissues, while Bacteroidetes has a lower abundance (
Figure 2A). To seek more details at the class level, we plotted the 3D-chord diagram to visualize the overlapping and diversity of classes within rice tissues. As a result, Gamma proteobacteria and Bacilli were more diversified and overlapped among rice tissues in contrast to Betaproteobacteria, Flavobacteria, and Sphingobacteria (
Figure 2B).
Figure 1. Rice endophytic bacterial communities. Each specified color in Ring-1 represents each phylum, and Ring-2 illustrates the source of bacterial communities from rice plants. An intermediated portion between Ring-1 and Ring-2 shows the plant-growth-promoting (PGP) attributes produced by rice endophytic bacterial strains; the (+) symbol indicates the presence of PGP traits, whereas the (−) sign indicates the absence of traits. The eight Arabic digits between in brackets indicate the accession number of the National Center for Biotechnology Information (NCBI) and references for taxonomic classification of bacterial communities (see the
Supplementary Materials Data 2 for high-resolution visualization of figure).
Figure 2. Relative abundance of rice endophytic bacteria diversity within rice tissues. (
A) Endophytic bacteria diversity at the phylum level; the isolation sources were plotted on the horizontal axis, and relative percentage of phyla is visualized by the vertical axis, each taxon showed with specified color in stacked columns. (
B) 3D chord diagram illustrates the rice endophytic bacteria diversity at the class level; each arc shows overlapping diversity within rice tissues; 3D visualization available on following link
https://beta.rstudioconnect.com/connect/#/apps/42c60c75-8ff2-4dce-ad48-c1446c8ccb55/access, accessed on 5 September 2021 Total Div.: presenting the total diversity within rice tissues (leaf, root, stem, shoot, seed, and including other sources). Other: including isolation sources of rice (grain filling, tillage stages, and also not-conformed sources).
The majority of the endophytic bacterial diversity was isolated from root portions that displayed more diverse communities. Many researchers isolated
Bacillus,
Pantoea,
Methylobacterium and
Sphingomonas genera, they are in high abundance with diverse species from the seed [
23,
24], root [
25,
26], and leaf part [
27,
28] of the rice plant (
Supplementary Materials Data 1, Figure S1). Most species of
Rhizobium,
Azospirillum,
Burkholderia, and
Herbaspirillum were isolated from root tissues with distinct species [
29,
30,
31,
32], and
Xanthomonas,
Flavobacterium, and
Knoellia genera were isolated from seeds (
Supplementary Materials Data 1, Figure S1) [
33,
34]. These endophytes genera also have been identified after isolation from different plants, such as ben tree, (
Moringa peregrina) [
35], maize (
Zea mays) [
36,
37],
Datura metel (local name Devil’s Trumpet) [
38], Kudouzi (
Sophora alopecuroides) [
39], Scots pine (
Pinus sylvestris) [
40], strawberry (
Fragaria ananassa) [
41,
42], wheat (
Triticum aestivum L.) [
43], rose gum (
Eucalyptus grandis) [
44], sugarcane (
Saccharum officinarum) [
45], grapevine (
Vitis vinifera) [
46,
47], poplar (
Populus deltoides) [
48], pepper (
Capsicum annuum) [
49], Huang-Qin (
Scutellaria baicalensis Georgi) [
50], cotton (
Gossypium hirsutum) [
51], potato (
Solanum tuberosum) [
52], cucumber (
Cucumis sativus) [
53], tobacco (
Nicotiana tabacum L.) [
54], peanut (
Echinopsis chamaecereus “Lutea”) [
55], tulasi (
Ocimum sanctum) [
56], and pea (
Pisum sativum L.) [
57]. Few studies have reported that bacterial genera are specific for seed colonization; for example, the
Pseudomonas putida mutant during the secretion-system investigations displayed the reduction in their potential to colonize with maize seeds [
58]. On the other hand, rice root endophytic bacteria are more diverse [
25,
59] compared to seed endophytes [
60,
61]; complete details can be visualized in
Figure 1.
Seeds can also be colonized by bacteria existing on the surfaces of flower petals, fruits, and stems [
62,
63], and by additional pollen grains’ bacteria [
64] that may be associated with ovules after pollination processes [
65]. Root endophytic bacteria can be explored by using the soil surrounding plant roots or plant litter [
36,
66]. Moreover, endophytic bacterial diversity variation is associated with physicochemical properties of soil, as well as atmospheric conditions, cropping history, geographic location, and other agricultural practices [
36,
67,
68]. In certain studies, it is suggested that root endophytic bacteria can mainly exist among the rhizosphere parts, and some of them can migrate through the xylem track to colonize the aerial parts [
69,
70].
Seed-borne pathogens can be persistent among new generations of plants. In the same way, seeds can also be harbored by endophytic bacteria that might be hereditary to new generations. Non-pathogenic bacteria have been categorized, from seeds and roots, parts of rice [
25,
29], maize [
37], wheat [
71], and other plants [
37,
72,
73]. Many endophytes responsible for plant-growth-promoting traits (such as IAA, ACC, siderophore production, nitrogen fixation, phosphorus production, zinc solubilization, and ammonia production) have been isolated from leaves, seeds, stems, and roots of rice plants (
Figure 1). These beneficial endophytes assist the host plants to maintain their growth and increase immunity against phytopathogens.
This entry is adapted from the peer-reviewed paper 10.3390/ijms221810165