Nasopharyngeal carcinoma (NPC) is one of the most common malignant tumours of the head and neck, and improving the efficiency of its diagnosis and treatment strategies is an important goal. With the development of the combination of artificial intelligence (AI) technology and medical imaging in recent years, an increasing number of studies have been conducted on image analysis of NPC using AI tools, especially radiomics and artificial neural network methods.
- nasopharyngeal carcinoma
- deep learning
- radiomics
- imaging
1. Introduction
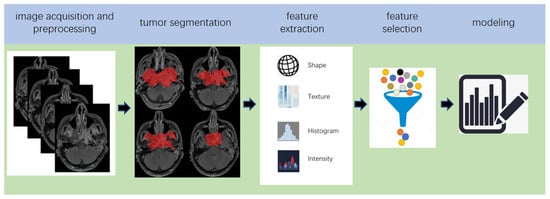
2. Pipeline of Radiomics
Radiomics, which was first proposed by Lambin in 2012 [7], is a relatively ‘young’ concept and is considered a natural extension of computer-aided diagnosis and detection systems [8]. It converts imaging data into a high-dimensional mineable feature space using a large number of automatically extracted data-characterization algorithms to reveal tumour features that may not be recognized by the naked eye and to quantitatively describe the tumour phenotype [9][10][11][12]. These extracted features are called radiomic features and include first-order statistics features, intensity histograms, shape- and size-based features, texture-based features, and wavelet features [13]. Conceptually speaking, radiomics belongs to the field of machine learning, although human participation is needed. The basic hypothesis of radiomics is that the constructed descriptive model (based on medical imaging data, sometimes supplemented by biological and/or medical data) can provide predictions of prognosis or diagnosis [14]. A radiomics study can be structured in five steps: data acquisition and pre-processing, tumour segmentation, feature extraction, feature selection, and modelling [15][16] (Figure 2).
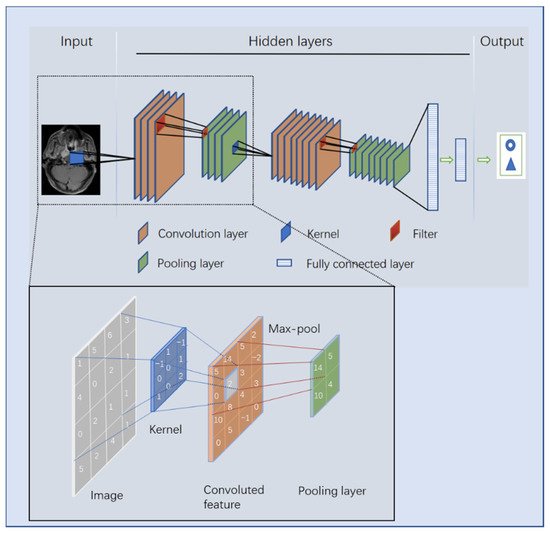
3. The Principle of DL
5. Studies Based on Radiomics
5.1. Prognosis Prediction
| Author, Year, Reference | Image | Sample Size (Patient) | Feature Selection | Modeling | Model Evaluation |
|---|---|---|---|---|---|
| Zhang, B. (2017) [13] | MRI | 108 | LASSO | CR, nomograms, calibration curves | C-index 0.776 |
| Zhang, B. (2017) [47] | MRI | 110 | L1-LOG, L1-SVM, RF, DC, EN-LOG, SIS | L2-LOG, KSVM, AdaBoost, LSVM, RF, Nnet, KNN, LDA, NB | AUC 0.846 |
| Zhang, B. (2017) [48] | MRI | 113 | LASSO | RS | AUC 0.886 |
| Ouyang, F.S. (2017) [49] | MRI | 100 | LASSO | RS | HR 7.28 |
| Lv, W. (2019) [50] | PET/CT | 128 | Univariate analysis with FDR, SC > 0.8 | CR | C-index 0.77 |
| Zhuo, E.H. (2019) [51] | MRI | 658 | Entropy-based consensus clustering method | SVM | C-index 0.814 |
| Zhang, L.L. (2019) [52] | MRI | 737 | RFE | CR and nomogram | C-index 0.73 |
| Yang, K. (2019) [53] | MRI | 224 | LASSO | CR and nomogram | C-index 0.811 |
| Ming, X. (2019) [54] | MRI | 303 | Non-negative matrix factorization | Chi-squared test, nomogram | C-index 0.845 |
| Zhang, L. (2019) [55] | MRI | 140 | LR-RFE | CR and nomogram | C-index 0.74 |
| Mao, J. (2019) [56] | MRI | 79 | Univariate analyses | CR | AUC 0.825 |
| Du, R. (2019) [57] | MRI | 277 | Hierarchal clustering analysis, PR | SVM | AUC 0.8 |
| Xu, H. (2020) [58] | PET/CT | 128 | Univariate CR, PR > 0.8 | CR | C-index 0.69 |
| Shen, H. (2020) [59] | MRI | 327 | LASSO, RFE | CR, RS | C-index 0.874 |
| Bologna, M. (2020) [60] | MRI | 136 | Intra-class correlation coefficient, SCC > 0.85 | CR | C-index 0.72 |
| Feng, Q. (2020) [61] | PET/MR | 100 | LASSO | CR | AUC 0.85 |
| Peng, L. (2021) [62] | PET/CT | 85 | W-test, Chi-square test, PR, RA | SFFS coupled with SVM | AUC 0.829 |
5.2. Assessment of Tumour Metastasis
The authors in [63] developed an MRI-based radiomics nomogram for the differential diagnosis of cervical spine lesions and metastasis after radiotherapy. A total of 279 radiomic features were extracted from the enhanced T1-weighted MRI, and eight radiomic features were selected using LASSO to establish a classifier model that obtained an AUC of 0.72 with the validation set.
In [64], the authors explored the issue of whether there was a difference between radiomic features derived from recurrent and non-recurrent regions within the tumour. Seven histogram features and 40 texture features were extracted from the MRI images of 14 patients with T4NxM0 NPC. The author proposed that there were seven features that were significantly different between the recurrent and non-recurrent regions.
In 2021, the study of [62], which was introduced in the section on prognosis prediction, established a model for the assessment of tumour metastasis simultaneously. The best AUC for predicting tumour metastasis was 0.829 (Table 2).
Table 2. Studies for assessing tumour metastasis using radiomics.
| Author, Year, Reference | Image | Sample Size | Feature Selection | Modeling | Model Evaluation |
|---|---|---|---|---|---|
| Zhang, L. (2019) [65] | MRI | 176 | LASSO | LR | AUC 0.792 |
| Zhong, X. (2020) [63] | MRI | 46 | LASSO | Nomogram | AUC 0.72 |
| Akram, F. (2020) [64] | MRI | 14 | Paired t-test and W-test | Shapiro-Wilk normality tests | p < 0.001 |
| Zhang, X. (2020) [66] | MRI | 238 | MRMR combined with 0.632 + bootstrap algorithms | RF | AUC 0.845 |
| Peng, L. (2021) [62] | PET/CT | 85 | W-test, PR, RA, Chi-square test | SFFS coupled with SVM | AUC 0.829 |
5.3. Tumour Diagnosis
Lv [67] established a diagnostic model to distinguish NPC from chronic nasopharyngitis using the logistic regression of leave-one-out cross-validation method. A total of 57 radiological features were extracted from the PET/CT of 106 patients, and AUCs between 0.81 and 0.89 were reported.
In [68], 76 patients were enrolled, including 41 with local recurrence and 35 with inflammation, as confirmed by pathology. A total of 487 radiomic features were extracted from the PET images. The performance was investigated for 42 cross-combinations derived from six feature selection methods and seven classifiers. The authors concluded that diagnostic models based on radiomic features showed higher AUCs (0.867–0.892) than traditional clinical indicators (AUC = 0.817).
5.4. Prediction of Therapeutic Effect
In [69], 108 patients with advanced NPC were included to establish the dataset. The ANOVA/Mann–Whitney U test, correlation analysis, and LASSO were used to select texture features, and multivariate logistic regression was used to establish a predictive model for the early response to neoadjuvant chemotherapy. Finally, an AUC of 0.905 was obtained for the validation cohort.
5.5. Predicting Complications
In [70], a radiomics model for predicting early acute xerostomia during radiation therapy was established based on CT images. Ridge CV and recursive feature elimination were used for feature selection, whereas linear regression was used for modelling. However, the study’s test cohort included only four patients with NPC and lacked sufficient evidence, despite the study reaching a precision of 0.922.
The authors in [71] established three radiomics models for the early diagnosis of radiation-induced temporal lobe injury based on the MRIs of 242 patients with NPC. The feature selection in the study was achieved by the Relief algorithm, which is different from other studies. The random forest algorithm was used to establish three early diagnosis models. The AUCs of the models in the test cohort were 0.830, 0.773, and 0.716, respectively.
6. Studies Based on DL
6.1. Prognosis Prediction
Yang [72] established a weakly-supervised, deep-learning network using an improved residual network (ResNet) with three input channels to achieve automated T staging of NPC. The images of multiple tumour layers of patients were labelled uniformly. The model output a predicted T-score for each slice and then selected the highest T-score slice for each patient to retrain the model to update the network weights. The accuracy of the model in the validation set was 75.59%, and the AUC was 0.943.
In [73], a DL model based on ResNet was established to predict the distant metastasis-free survival of locally advanced NPC. In contrast to the studies published in 2020, the authors of this study removed the background noise and segmented the tumour region as the input image of the DL network. Finally, the optimal AUC of the multiple models combined with the clinical features was 0.808 (Table 7).
6.2. Image Synthesis
6.3. Detection and/or Diagnosis
Two similar studies, [76][77], based on pathological images were conducted. The authors in [76] used 1970 whole slide pathological images of 731 cases: 316 cases of inflammation, 138 cases of lymphoid hyperplasia, and 277 cases of NPC. The second study used 726 nasopharyngeal biopsies consisting of 363 images of NPC and 363 of benign nasopharyngeal tissue [77]. In [76], Inception-v3 was used to build the classifier, while ResNeXt, a deep neural network with a residual and inception architecture, was used to build the classifier in [77]. The AUCs obtained in [76][77] were 0.936 and 0.985, respectively.
6.4. Segmentation
7. Deep Learning-Based Radiomics
DL has shown great potential to dominate the field of image analysis. In ROI [80] and feature extraction tasks [81][82], which lay in the implementation pipeline of radiomics, DL has achieved good results. After completing the model training, DL can automatically analyse images, which is one of the greatest strengths compared to radiomics. Many researchers have introduced DL into radiomics (termed deep learning-based radiomics, DLR) and achieved encouraging results [83]. This may be a trend for the application of AI tools in medical imaging in the future.
7.1. Studies Based on Deep Learning-Based Radiomics (DLR)
In [84], Zhang innovatively combined the clinical features of patients with nasopharyngeal cancer, the radiomic features based on MRIs, and the DCNN model based on pathological images to construct a multi-scale nomogram to predict the failure-free survival of patients with NPC. The nomogram showed a consistent significant improvement for predicting treatment failure compared with the clinical model in the internal test (C-index: 0.828 vs. 0.602, p < 0.050) and external test (C-index: 0.834 vs. 0.679, p < 0.050) cohorts. (Table 11)
8. Future Work
Research on radiomics and DL in NPC imaging has only started in recent years. Therefore, there are still many issues that need further research in the future: linking NPC imaging features with tumour genes/molecules to promote the development of precision medicine for non-invasive, rapid, and low-cost approaches; using multi-stage dynamic imaging to assess tumour response to drugs/radiotherapy and predict the risk of radiation therapy in surrounding vital organs to guide treatment decisions; and bridging the gap from the AI tools established in studies to clinical applications. In addition, current studies based on nasal endoscopic images and pathological images are lacking. In particular, accurate and rapid screening of NPC is of great significance, considering that endoscopic images are usually the primary screening images for most patients. Further high-quality research in this regard is needed. Finally, there is still a lack of large-scale, comprehensive, and fully labelled datasets for NPC; datasets similar to those that are available for lung and brain tumours. The establishment of large-scale public datasets is an important task in the future.
This entry is adapted from the peer-reviewed paper 10.3390/diagnostics11091523
References
- Chen, Y.; Chan, A.T.C.; Le, Q.-T.; Blanchard, P.; Sun, Y.; Ma, J. Nasopharyngeal carcinoma. Lancet 2019, 394, 64–80.
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020; Available online: https://gco.iarc.fr/today (accessed on 31 March 2021).
- Butterfield, D. Impacts of Water and Export Market Restrictions on Palestinian Agriculture. Toronto: McMaster University and Econometric Research Limited, Applied Research Institute of Jerusalem (ARIJ). Available online: http://www.socserv.mcmaster.ca/kubursi/ebooks/water.htm (accessed on 31 March 2021).
- Chua, M.L.K.; Wee, J.T.S.; Hui, E.P.; Chan, A.T.C. Nasopharyngeal carcinoma. Lancet 2016, 387, 1012–1024.
- Yuan, H.; Lai-Wan, K.D.; Kwong, D.L.-W.; Fong, D.; King, A.; Vardhanabhuti, V.; Lee, V.; Khong, P.-L. Cervical nodal volume for prognostication and risk stratification of patients with nasopharyngeal carcinoma, and implications on the TNM-staging system. Sci. Rep. 2017, 7, 10387.
- Chan, A.T.C.; Grégoire, V.; Lefebvre, J.L.; Licitra, L.; Hui, E.P.; Leung, S.F.; Felip, E. Nasopharyngeal cancer: EHNS-ESMO-ESTRO clinical practice guidelines for diagnosis, treatment andfollow-up. Ann. Oncol. 2012, 23, vii83–vii85.
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446.
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577.
- Aerts, H.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Data from: Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006.
- Yip, S.S.F.; Aerts, H.J.W.L. Applications and limitations of radiomics. Phys. Med. Biol. 2016, 61, R150–R166.
- Rahmim, A.; Schmidtlein, C.R.; Jackson, A.; Sheikhbahaei, S.; Marcus, C.; Ashrafinia, S.; Soltani, M.; Subramaniam, R.M. A novel metric for quantification of homogeneous and heterogeneous tumors in PET for enhanced clinical outcome prediction. Phys. Med. Biol. 2015, 61, 227–242.
- Buvat, I.; Orlhac, F.; Soussan, M.; Orhlac, F. Tumor Texture Analysis in PET: Where Do We Stand? J. Nucl. Med. 2015, 56, 1642–1644.
- Zhang, B.; Tian, J.; Dong, D.; Gu, D.; Dong, Y.; Zhang, L.; Lian, Z.; Liu, J.; Luo, X.; Pei, S.; et al. Radiomics Features of Multiparametric MRI as Novel Prognostic Factors in Advanced Nasopharyngeal Carcinoma. Clin. Cancer Res. 2017, 23, 4259–4269.
- Afshar, P.; Mohammadi, A.; Plataniotis, K.; Oikonomou, A.; Benali, H. From Handcrafted to Deep-Learning-Based Cancer Radiomics: Challenges and Opportunities. IEEE Signal Process. Mag. 2019, 36, 132–160.
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762.
- Liu, Z.; Wang, S.; Dong, D.; Wei, J.; Fang, C.; Zhou, X.; Sun, K.; Li, L.; Li, B.; Wang, M.; et al. The Applications of Radiomics in Precision Diagnosis and Treatment of Oncology: Opportunities and Challenges. Theranostics 2019, 9, 1303–1322.
- Ford, J.; Dogan, N.; Young, L.; Yang, F. Quantitative Radiomics: Impact of Pulse Sequence Parameter Selection on MRI-Based Textural Features of the Brain. Contrast Media Mol. Imaging 2018, 2018.
- Peng, H.; Dong, D.; Fang, M.-J.; Li, L.; Tang, L.-L.; Chen, L.; Li, W.-F.; Mao, Y.-P.; Fan, W.; Liu, L.-Z.; et al. Prognostic Value of Deep Learning PET/CT-Based Radiomics: Potential Role for Future Individual Induction Chemotherapy in Advanced Nasopharyngeal Carcinoma. Clin. Cancer Res. 2019, 25, 4271–4279.
- Guo, Y.; Hu, Y.; Qiao, M.; Wang, Y.; Yu, J.; Li, J.; Chang, C. Radiomics Analysis on Ultrasound for Prediction of Biologic Behavior in Breast Invasive Ductal Carcinoma. Clin. Breast Cancer 2018, 18, e335–e344.
- Song, G.; Xue, F.; Zhang, C. A Model Using Texture Features to Differentiate the Nature of Thyroid Nodules on Sonography. J. Ultrasound Med. 2015, 34, 1753–1760.
- Polan, D.F.; Brady, S.L.; Kaufman, R.A. Tissue segmentation of computed tomography images using a Random Forest algorithm: A feasibility study. Phys. Med. Biol. 2016, 61, 6553.
- Hatt, M.; Tixier, F.; Pierce, L.; Kinahan, P.; Le Rest, C.C.; Visvikis, D. Characterization of PET/CT images using texture analysis: The past, the present… any future? Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 151–165.
- Zhang, L.; Fried, D.V.; Fave, X.J.; Hunter, L.A.; Yang, J.; Court, L.E. IBEX: An open infrastructure software platform to facilitate collaborative work in radiomics. Med. Phys. 2015, 42, 1341–1353.
- Limkin, E.J.; Sun, R.; Dercle, L.; Zacharaki, E.I.; Robert, C.; Reuzé, S.; Schernberg, A.; Paragios, N.; Deutsch, E.; Ferté, C. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann. Oncol. 2017, 28, 1191–1206.
- Zhang, Y.; Oikonomou, A.; Wong, A.; Haider, M.A.; Khalvati, F. Radiomics-based Prognosis Analysis for Non-Small Cell Lung Cancer. Sci. Rep. 2017, 7, srep46349.
- Grootjans, W.; Tixier, F.; van der Vos, C.S.; Vriens, D.; Le Rest, C.C.; Bussink, J.; Oyen, W.J.; de Geus-Oei, L.-F.; Visvikis, D.; Visser, E.P. The Impact of Optimal Respiratory Gating and Image Noise on Evaluation of Intratumor Heterogeneity on 18F-FDG PET Imaging of Lung Cancer. J. Nucl. Med. 2016, 57, 1692–1698.
- Santos, M.K.; Júnior, J.R.F.; Wada, D.T.; Tenório, A.P.M.; Barbosa, M.H.N.; Marques, P.M.D.A. Artificial intelligence, machine learning, computer-aided diagnosis, and radiomics: Advances in imaging towards to precision medicine. Radiol. Bras. 2019, 52, 387–396.
- Hamet, P.; Tremblay, J. Artificial intelligence in medicine. Metabolism 2017, 69, S36–S40.
- Moor, J. The Dartmouth College artificial intelligence conference: The next fifty years. Ai Mag. 2006, 27, 87.
- Zhou, X.; Li, C.; Rahaman, M.; Yao, Y.; Ai, S.; Sun, C.; Wang, Q.; Zhang, Y.; Li, M.; Li, X.; et al. A Comprehensive Review for Breast Histopathology Image Analysis Using Classical and Deep Neural Networks. IEEE Access 2020, 8, 90931–90956.
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine Learning for Medical Imaging. Radiographics 2017, 37, 505–515.
- Foster, K.R.; Koprowski, R.; Skufca, J.D. Machine learning, medical diagnosis, and biomedical engineering research—Commentary. Biomed. Eng. Online 2014, 13, 94.
- Rashidi, H.H.; Tran, N.K.; Betts, E.V.; Howell, L.P.; Green, R. Artificial intelligence and machine learning in pathology: The present landscape of supervised methods. Acad. Pathol. 2019, 6.
- Jafari, M.; Wang, Y.; Amiryousefi, A.; Tang, J. Unsupervised Learning and Multipartite Network Models: A Promising Approach for Understanding Traditional Medicine. Front. Pharmacol. 2020, 11, 1319.
- Bzdok, D.; Krzywinski, M.; Altman, N. Machine learning: Supervised methods. Nat. Methods 2018, 15, 5–6.
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical Image Analysis using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226.
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444.
- Zipser, D.; Andersen, R.A. A back-propagation programmed network that simulates response properties of a subset of posterior parietal neurons. Nat. Cell Biol. 1988, 331, 679–684.
- Kriegeskorte, N.; Golan, T. Neural network models and deep learning. Curr. Biol. 2019, 29, R231–R236.
- Manisha; Dhull, S.K.; Singh, K.K. ECG Beat Classifiers: A Journey from ANN To DNN. Procedia Comput. Sci. 2020, 167, 747–759.
- Soffer, S.; Ben-Cohen, A.; Shimon, O.; Amitai, M.M.; Greenspan, H.; Klang, E. Convolutional Neural Networks for Radiologic Images: A Radiologist’s Guide. Radiology 2019, 290, 590–606.
- Deng, Y.; Bao, F.; Dai, Q.; Wu, L.F.; Altschuler, S.J. Scalable analysis of cell-type composition from single-cell transcriptomics using deep recurrent learning. Nat. Methods 2019, 16, 311–314.
- Li, Y.; Ma, X.; Zhou, X.; Cheng, P.; He, K.; Li, C. Knowledge Enhanced LSTM for Coreference Resolution on Biomedical Texts. Bioinformatics 2021.
- Hua, Y.; Guo, J.; Zhao, H. Deep belief networks and deep learning. In Proceedings of the 2015 International Conference on Intelligent Computing and Internet of Things, Harbin, China, 17–18 January 2015; pp. 1–4.
- Ko, W.Y.; Siontis, K.C.; Attia, Z.I.; Carter, R.E.; Kapa, S.; Ommen, S.R.; Demuth, S.J.; Ackerman, M.J.; Gersh, B.J.; Arruda-Olson, A.M.; et al. Detection of hypertrophic cardiomyopathy using a convolutional neural network-enabled electrocardiogram. J. Am. Coll. Cardiol. 2020, 75, 722–733.
- Skrede, O.-J.; De Raedt, S.; Kleppe, A.; Hveem, T.S.; Liestøl, K.; Maddison, J.; Askautrud, H.; Pradhan, M.; Nesheim, J.A.; Albregtsen, F.; et al. Deep learning for prediction of colorectal cancer outcome: A discovery and validation study. Lancet 2020, 395, 350–360.
- Zhang, B.; He, X.; Ouyang, F.; Gu, D.; Dong, Y.; Zhang, L.; Mo, X.; Huang, W.; Tian, J.; Zhang, S. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett. 2017, 403, 21–27.
- Zhang, B.; Ouyang, F.; Gu, D.; Dongsheng, G.; Zhang, L.; Mo, X.; Huang, W.; Zhang, S. Advanced nasopharyngeal carcinoma: Pre-treatment prediction of progression based on multi-parametric MRI radiomics. Oncotarget 2017, 8, 72457–72465.
- Ouyang, F.S.; Guo, B.L.; Zhang, B.; Dong, Y.H.; Zhang, L.; Mo, X.K.; Huang, W.H.; Zhang, S.X.; Hu, Q.G. Exploration and validation of radiomics signature as an independent prognostic biomarker in stage III-IVb Nasopharyngeal carcinoma. Oncotarget 2017, 8, 74869–74879.
- Lv, W.; Yuan, Q.; Wang, Q.; Ma, J.; Feng, Q.; Chen, W.; Rahmim, A.; Lu, L. Radiomics Analysis of PET and CT Components of PET/CT Imaging Integrated with Clinical Parameters: Application to Prognosis for Nasopharyngeal Carcinoma. Mol. Imaging Biol. 2019, 21, 954–964.
- Zhuo, E.-H.; Zhang, W.-J.; Li, H.-J.; Zhang, G.-Y.; Jing, B.-Z.; Zhou, J.; Cui, C.-Y.; Chen, M.-Y.; Sun, Y.; Liu, L.-Z.; et al. Radiomics on multi-modalities MR sequences can subtype patients with non-metastatic nasopharyngeal carcinoma (NPC) into distinct survival subgroups. Eur. Radiol. 2019, 29, 5590–5599.
- Zhang, L.L.; Huang, M.Y.; Li, Y.; Liang, J.H.; Gao, T.S.; Deng, B.; Yao, J.J.; Lin, L.; Chen, F.P.; Huang, X.D.; et al. Pretreatment MRI radiomics analysis allows for reliable prediction of local recurrence in non-metastatic T4 Nasopharyngeal carcinoma. EBioMedicine 2019, 42, 270–280.
- Yang, K.; Tian, J.; Zhang, B.; Li, M.; Xie, W.; Zou, Y.; Tan, Q.; Liu, L.; Zhu, J.; Shou, A.; et al. A multidimensional nomogram combining overall stage, dose volume histogram parameters and radiomics to predict progression-free survival in patients with locoregionally advanced nasopharyngeal carcinoma. Oral Oncol. 2019, 98, 85–91.
- Ming, X.; Oei, R.W.; Zhai, R.; Kong, F.; Du, C.; Hu, C.; Hu, W.; Zhang, Z.; Ying, H.; Wang, J. MRI-based radiomics signature is a quantitative prognostic biomarker for nasopharyngeal carcinoma. Sci. Rep. 2019, 9, 10412.
- Zhang, L.; Zhou, H.; Gu, D.; Tian, J.; Zhang, B.; Dong, D.; Mo, X.; Liu, J.; Luo, X.; Pei, S.; et al. Radiomic Nomogram: Pretreatment Evaluation of Local Recurrence in Nasopharyngeal Carcinoma based on MR Imaging. J. Cancer 2019, 10, 4217–4225.
- Mao, J.; Fang, J.; Duan, X.; Yang, Z.; Cao, M.; Zhang, F.; Lu, L.; Zhang, X.; Wu, X.; Ding, Y.; et al. Predictive value of pretreatment MRI texture analysis in patients with primary nasopharyngeal carcinoma. Eur. Radiol. 2019, 29, 4105–4113.
- Du, R.; Lee, V.H.; Yuan, H.; Lam, K.-O.; Pang, H.H.; Chen, Y.; Lam, E.Y.; Khong, P.-L.; Lee, A.W.; Kwong, D.L.; et al. Radiomics Model to Predict Early Progression of Nonmetastatic Nasopharyngeal Carcinoma after Intensity Modulation Radiation Therapy: A Multicenter Study. Radiol. Artif. Intell. 2019, 1, e180075.
- Xu, H.; Lv, W.; Feng, H.; Du, D.; Yuan, Q.; Wang, Q.; Dai, Z.; Yang, W.; Feng, Q.; Ma, J.; et al. Subregional Radiomics Analysis of PET/CT Imaging with Intratumor Partitioning: Application to Prognosis for Nasopharyngeal Carcinoma. Mol. Imaging Biol. 2020, 22, 1414–1426.
- Shen, H.; Wang, Y.; Liu, D.; Lv, R.; Huang, Y.; Peng, C.; Jiang, S.; Wang, Y.; He, Y.; Lan, X.; et al. Predicting Progression-Free Survival Using MRI-Based Radiomics for Patients with Nonmetastatic Nasopharyngeal Carcinoma. Front. Oncol. 2020, 10, 618.
- Bologna, M.; Corino, V.; Calareso, G.; Tenconi, C.; Alfieri, S.; Iacovelli, N.A.; Cavallo, A.; Cavalieri, S.; Locati, L.; Bossi, P.; et al. Baseline MRI-Radiomics Can Predict Overall Survival in Non-Endemic EBV-Related Nasopharyngeal Carcinoma Patients. Cancers 2020, 12, 2958.
- Feng, Q.; Liang, J.; Wang, L.; Niu, J.; Ge, X.; Pang, P.; Ding, Z. Radiomics Analysis and Correlation with Metabolic Parameters in Nasopharyngeal Carcinoma Based on PET/MR Imaging. Front. Oncol. 2020, 10, 1619.
- Peng, L.; Hong, X.; Yuan, Q.; Lu, L.; Wang, Q.; Chen, W. Prediction of local recurrence and distant metastasis using radiomics analysis of pretreatment nasopharyngeal FDG PET/CT images. Ann. Nucl. Med. 2021, 35, 458–468.
- Zhong, X.; Li, L.; Jiang, H.; Yin, J.; Lu, B.; Han, W.; Li, J.; Zhang, J. Cervical spine osteoradionecrosis or bone metastasis after radiotherapy for nasopharyngeal carcinoma? The MRI-based radiomics for characterization. BMC Med. Imaging 2020, 20, 104.
- Akram, F.; Koh, P.E.; Wang, F.; Zhou, S.; Tan, S.H.; Paknezhad, M.; Park, S.; Hennedige, T.; Thng, C.H.; Lee, H.K.; et al. Exploring MRI based radiomics analysis of intratumoral spatial heterogeneity in locally advanced nasopharyngeal carcinoma treated with intensity modulated radiotherapy. PLoS ONE 2020, 15, e0240043.
- Zhang, L.; Dong, D.; Li, H.; Tian, J.; Ouyang, F.; Mo, X.; Zhang, B.; Luo, X.; Lian, Z.; Pei, S.; et al. Development and validation of a magnetic resonance imaging-based model for the prediction of distant metastasis before initial treatment of nasopharyngeal carcinoma: A retrospective cohort study. EBioMedicine 2019, 40, 327–335.
- Zhang, X.; Zhong, L.; Zhang, B.; Zhang, L.; Du, H.; Lu, L.; Zhang, S.; Yang, W.; Feng, Q. The effects of volume of interest delineation on MRI-based radiomics analysis: Evaluation with two disease groups. Cancer Imaging 2019, 19, 89–112.
- Lv, W.; Yuan, Q.; Wang, Q.; Ma, J.; Jiang, J.; Yang, W.; Feng, Q.; Chen, W.; Rahmim, A.; Lu, L. Robustness versus disease differentiation when varying parameter settings in radiomics features: Application to nasopharyngeal PET/CT. Eur. Radiol. 2018, 28, 3245–3254.
- Du, D.; Feng, H.; Lv, W.; Ashrafinia, S.; Yuan, Q.; Wang, Q.; Yang, W.; Feng, Q.; Chen, W.; Rahmim, A.; et al. Machine Learning Methods for Optimal Radiomics-Based Differentiation Between Recurrence and Inflammation: Application to Nasopharyngeal Carcinoma Post-therapy PET/CT Images. Mol. Imaging Biol. 2020, 22, 730–738.
- Yongfeng, P.; Chuner, J.; Lei, W.; Fengqin, Y.; Zhimin, Y.; Zhenfu, F.; Haitao, J.; Yangming, J.; Fangzheng, W. The usefulness of pre-treatment MR-based radiomics on early response of neoadjuvant chemotherapy in patients with locally advanced Nasopharyngeal carcinoma. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2021, 28, 605–613.
- Liu, Y.; Shi, H.; Huang, S.; Chen, X.; Zhou, H.; Chang, H.; Xia, Y.; Wang, G.; Yang, X. Early prediction of acute xerostomia during radiation therapy for nasopharyngeal cancer based on delta radiomics from CT images. Quant. Imaging Med. Surg. 2019, 9, 1288–1302.
- Zhang, B.; Lian, Z.; Zhong, L.; Zhang, X.; Dong, Y.; Chen, Q.; Zhang, L.; Mo, X.; Huang, W.; Yang, W.; et al. Machine-learning based MRI radiomics models for early detection of radiation-induced brain injury in nasopharyngeal carcinoma. BMC Cancer 2020, 20, 502.
- Yang, Q.; Guo, Y.; Ou, X.; Wang, J.; Hu, C. Automatic T Staging Using Weakly Supervised Deep Learning for Nasopharyngeal Carcinoma on MR Images. J. Magn. Reson. Imaging 2020, 52, 1074–1082.
- Zhang, L.; Wu, X.; Liu, J.; Zhang, B.; Mo, X.; Chen, Q.; Fang, J.; Wang, F.; Li, M.; Chen, Z.; et al. MRI-based deep-learning model for distant metastasis-free survival in locoregionally advanced Nasopharyngeal carcinoma. J. Magn. Reson. Imaging 2021, 53, 167–178.
- Tie, X.; Lam, S.K.; Zhang, Y.; Lee, K.H.; Au, K.H.; Cai, J. Pseudo-CT generation from multi-parametric MRI using a novel multi-channel multi-path conditional generative adversarial network for Nasopharyngeal carcinoma patients. Med. Phys. 2020, 47, 1750–1762.
- Peng, Y.; Chen, S.; Qin, A.; Chen, M.; Gao, X.; Liu, Y.; Miao, J.; Gu, H.; Zhao, C.; Deng, X.; et al. Magnetic resonance-based synthetic computed tomography images generated using generative adversarial networks for nasopharyngeal carcinoma radiotherapy treatment planning. Radiother. Oncol. 2020, 150, 217–224.
- Diao, S.; Hou, J.; Yu, H.; Zhao, X.; Sun, Y.; Lambo, R.L.; Xie, Y.; Liu, L.; Qin, W.; Luo, W. Computer-Aided Pathologic Diagnosis of Nasopharyngeal Carcinoma Based on Deep Learning. Am. J. Pathol. 2020, 190, 1691–1700.
- Chuang, W.-Y.; Chang, S.-H.; Yu, W.-H.; Yang, C.-K.; Yeh, C.-J.; Ueng, S.-H.; Liu, Y.-J.; Chen, T.-D.; Chen, K.-H.; Hsieh, Y.-Y.; et al. Successful Identification of Nasopharyngeal Carcinoma in Nasopharyngeal Biopsies Using Deep Learning. Cancers 2020, 12, 507.
- Li, S.; Xiao, J.; He, L.; Peng, X.; Yuan, X. The Tumor Target Segmentation of Nasopharyngeal Cancer in CT Images Based on Deep Learning Methods. Technol. Cancer Res. Treat. 2019, 18.
- Bai, X.; Hu, Y.; Gong, G.; Yin, Y.; Xia, Y. A deep learning approach to segmentation of nasopharyngeal carcinoma using computed tomography. Biomed. Signal. Process. Control. 2021, 64, 102246.
- Shboul, Z.; Alam, M.; Vidyaratne, L.; Pei, L.; Elbakary, M.I.; Iftekharuddin, K.M. Feature-Guided Deep Radiomics for Glioblastoma Patient Survival Prediction. Front. Neurosci. 2019, 13, 966.
- Paul, R.; Hawkins, S.H.; Schabath, M.B.; Gillies, R.J.; Hall, L.O.; Goldgof, D.B. Predicting malignant nodules by fusing deep features with classical radiomics features. J. Med. Imaging 2018, 5, 011021.
- Bizzego, A.; Bussola, N.; Salvalai, D.; Chierici, M.; Maggio, V.; Jurman, G.; Furlanello, C. Integrating deep and radiomics features in cancer bioimaging. In Proceedings of the 2019 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Certosa di Pontignano, Siena–Tuscany, Italy, 9–11 July 2019; pp. 1–8.
- Hatt, M.; Parmar, C.; Qi, J.; El Naqa, I. Machine (Deep) Learning Methods for Image Processing and Radiomics. IEEE Trans. Radiat. Plasma Med. Sci. 2019, 3, 104–108.
- Zhang, F.; Zhong, L.-Z.; Zhao, X.; Dong, D.; Yao, J.-J.; Wang, S.-Y.; Liu, Y.; Zhu, D.; Wang, Y.; Wang, G.-J.; et al. A deep-learning-based prognostic nomogram integrating microscopic digital pathology and macroscopic magnetic resonance images in nasopharyngeal carcinoma: A multi-cohort study. Ther. Adv. Med. Oncol. 2020, 12.
