As widely acknowledged, 40–50% of all melanoma patients harbour an activating BRAF mutation (mostly BRAF V600E). The identification of the RAS–RAF–MEK–ERK (MAP kinase) signalling pathway and its targeting has represented a valuable milestone for the advanced and, more recently, for the completely resected stage III and IV melanoma therapy management. However, despite progress in BRAF-mutant melanoma treatment, the two different approaches approved so far for metastatic disease, immunotherapy and BRAF+MEK inhibitors, allow a 5-year survival of no more than 60%, and most patients relapse during treatment due to acquired mechanisms of resistance. Deep insight into BRAF gene biology is fundamental to describe the acquired resistance mechanisms (primary and secondary) and to understand the molecular pathways that are now being investigated in preclinical and clinical studies with the aim of improving outcomes in BRAF-mutant patients.
- melanoma
- BRAF mutation
- immunotherapy
- targeted therapy
1. Introduction
2. BRAF Gene Biology
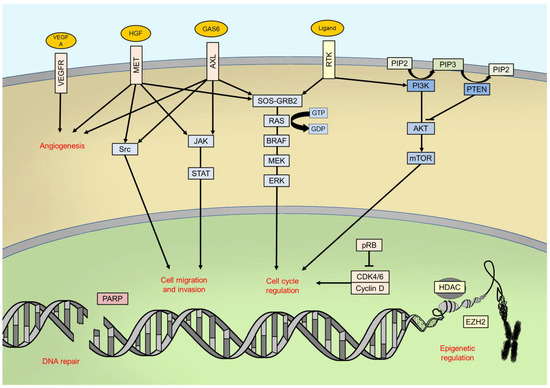
3. BRAF Mutations in Melanoma: Epidemiology and Clinic-Pathological Correlations
3.1. BRAF V600 Mutations
| Mutation | Frequency (among BRAF V600 Mutations) | DFS | PFS (with TT) | TMB | Response to ICIs |
|---|---|---|---|---|---|
| V600E | 70–88% | Longer | Longer | Lower | Good |
| V600K | 10–20% | Shorter | Shorter | Higher | Better |
3.2. BRAF Non-V600 Mutations
This entry is adapted from the peer-reviewed paper 10.3390/ijms22073474
References
- Palumbo, G.; Di Lorenzo, G.; Ottaviano, M.; Damiano, V. The future of melanoma therapy: Developing new drugs and improving the use of old ones. Future Oncol. 2016, 12, 2531–2534.
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954.
- Funck-Brentano, E.; Malissen, N.; Roger, A.; Lebbé, C.; Deilhes, F.; Frénard, C.; Dréno, B.; Meyer, N.; Grob, J.J.; Tétu, P.; et al. Which adjuvant treatment for patients with BRAFV600-mutant cutaneous melanoma? In Annales de Dermatologie et de Vénéréologie; Cribier, B., Ed.; Elsevier: Amsterdam, The Netherlands, 2021.
- Giunta, E.F.; De Falco, V.; Napolitano, S.; Argenziano, G.; Brancaccio, G.; Moscarella, E.; Ciardiello, D.; Ciardiello, F.; Troiani, T. Optimal treatment strategy for metastatic melanoma patients harboring BRAF-V600 mutations. Ther. Adv. Med. Oncol. 2020, 12, 1758835920925219.
- Ottaviano, M.; De Placido, S.; Ascierto, P.A. Recent success and limitations of immune checkpoint inhibitors for cancer: A lesson from melanoma. Virchows Arch. 2019, 474, 421–432.
- Peyssonnaux, C.; Eychene, A. The Raf/MEK/ERK pathway: New concepts of activation. Biol. Cell 2001, 93, 53–62.
- Morrison, D.K. Map kinase pathways. Cold Spring Harb. Perspect. Biol. 2012, 4, a011254.
- Cuevas, B.D.; Abell, A.N.; Johnson, G.L. Role of mitogen-activated protein kinase kinase kinases in signal integration. Oncogene 2007, 26, 3159–3171.
- Aramini, J.M.; Vorobiev, S.M.; Tuberty, L.M.; Janjua, H.; Campbell, E.T.; Seetharaman, J.; Su, M.; Huang, Y.J.; Acton, T.B.; Xiao, R.; et al. The RAS-binding domain of human BRAF protein serine/threonine kinase exhibits allosteric conformational changes upon binding HRAS. Structure 2015, 23, 1382–1393.
- Park, E.; Rawson, S.; Li, K.; Kim, B.W.; Ficarro, S.B.; Pino, G.G.; Sharif, H.; Marto, J.A.; Jeon, H.; Eck, M.J. Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes. Nature 2010, 575, 545–550.
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.; Chang, F.; Lehmann, B.; Terrian, D.M.; Milella, M.; Tafuri, A.; et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta 2007, 1773, 1263–1284.
- Steelman, L.S.; Pohnert, S.C.; Shelton, J.G.; Franklin, R.A.; Bertrand, F.E.; McCubrey, J.A. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia 2004, 18, 189–218.
- Wan, P.T.; Garnett, M.J.; Roe, S.M.; Lee, S.; Niculescu-Duvaz, D.; Good, V.M.; Jones, C.M.; Marshall, C.J.; Springer, C.J.; Barford, D.; et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004, 116, 855–867.
- Johnson, D.B.; Dahlman, K.B. Class matters: Sensitivity of BRAF-mutant melanoma to MAPK inhibition. Clin. Cancer Res. 2018, 24, 6107–6109.
- Long, G.V.; Menzies, A.M.; Nagrial, A.M.; Haydu, L.E.; Hamilton, A.L.; Mann, G.J.; Hughes, T.M.; Thompson, J.F.; Scolyer, R.A.; Kefford, R.F. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J. Clin. Oncol. 2011, 29, 1239–1246.
- Bauer, J.; Buttner, P.; Murali, R.; Okamoto, I.; Kolaitis, N.A.; Landi, M.T.; Scolyer, R.A.; Bastian, B.C. BRAF mutations in cutaneous melanoma are independently associated with age, anatomic site of the primary tumor, and the degree of solar elastosis at the primary tumor site. Pigment Cell Melanoma Res. 2011, 24, 345–351.
- Gopal, P.; Sarihan, E.I.; Chie, E.K.; Kuzmishin, G.; Doken, S.; Pennell, N.A.; Raymond, D.P.; Murthy, S.C.; Ahmad, U.; Raja, S.; et al. Clonal selection confers distinct evolutionary trajectories in BRAF-driven cancers. Nat. Commun. 2019, 10, 5143.
- Rubinstein, J.C.; Sznol, M.; Pavlick, A.C.; Ariyan, S.; Cheng, E.; Bacchiocchi, A.; Kluger, H.M.; Narayan, D.; Halaban, R. Incidence of the V600K mutation among melanoma patients with BRAF mutations, and potential therapeutic response to the specific BRAF inhibitor PLX4032. J. Transl. Med. 2010, 8, 67.
- Heinzerling, L.; Kühnapfel, S.; Meckbach, D.; Baiter, M.; Kaempgen, E.; Keikavoussi, P.; Schuler, G.; Agaimy, A.; Bauer, J.; Hartmann, A.; et al. Rare BRAF mutations in melanoma patients: Implications for molecular testing in clinical practice. Br. J. Cancer 2013, 108, 2164–2171.
- Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 2015, 161, 1681–1696.
- Menzies, A.M.; Haydu, L.E.; Visintin, L.; Carlino, M.S.; Howle, J.R.; Thompson, J.F.; Kefford, R.F.; Scolyer, R.A.; Long, G.V. Distinguishing clinicopathologic features of patients with V600E and V600K BRAF-mutant metastatic melanoma. Clin. Cancer Res. 2012, 18, 3242–3249.
- Pires da Silva, I.; Wang, K.Y.X.; Wilmott, J.S.; Holst, J.; Carlino, M.S.; Park, J.J.; Quek, C.; Wongchenko, M.; Yan, Y.; Mann, G.; et al. Distinct molecular profiles and immunotherapy treatment outcomes of V600E and V600K BRAF-mutant melanoma. Clin. Cancer Res. 2019, 25, 1272–1279.
- Li, Y.; Umbach, D.M.; Li, L. Putative genomic characteristics of BRAF V600K versus V600E cutaneous melanoma. Melanoma Res. 2017, 27, 527–535.
- Ny, L.; Hernberg, M.; Nyakas, M.; Koivunen, J.; Oddershede, L.; Yoon, M.; Wang, X.; Guyot, P.; Geisler, J. BRAF mutational status as a prognostic marker for survival in malignant melanoma: A systematic review and meta-analysis. Acta Oncol. 2020, 59, 833–844.
- Adler, N.R.; Wolfe, R.; Kelly, J.W.; Haydon, A.; McArthur, G.A.; McLean, C.A.; Mar, V.J. Tumour mutation status and sites of metastasis in patients with cutaneous melanoma. Br. J. Cancer 2017, 117, 1026–1035.
- Menzer, C.; Menzies, A.M.; Carlino, M.S.; Reijers, I.; Groen, E.J.; Eigentler, T.; de Groot, J.W.B.; van der Veldt, A.A.M.; Johnson, D.B.; Meiss, F.; et al. Targeted therapy in advanced melanoma with rare BRAF mutations. J. Clin. Oncol. 2019, 37, 3142–3151.
- Dahlman, K.B.; Xia, J.; Hutchinson, K.; Jia, P.; Atefi, M.; Su, Z.; Branch, S.; Lyle, P.L.; Hicks, D.J.; Bozon, V.; et al. BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov. 2012, 2, 791–797.
- Zheng, G.; Tseng, L.; Chen, G.; Haley, L.; Illei, P.; Gocke, C.D.; Eshleman, J.R.; Lin, M.T. Clinical detection and categorization of uncommon and concomitant mutations involving BRAF. BMC Cancer 2015, 15, 779.
- Yao, Z.; Yaeger, R.; Rodrik-Outmezguine, V.S.; Tao, A.; Torres, N.M.; Chang, M.T.; Drosten, M.; Zhao, H.; Cecchi, F.; Hembrough, T.; et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017, 548, 234–238.
- Wu, X.; Yan, J.; Dai, J.; Ma, M.; Tang, H.; Yu, J.; Xu, T.; Yu, H.; Si, L.; Chi, Z.; et al. Mutations in BRAF codons 594 and 596 predict good prognosis in melanoma. Oncol. Lett. 2017, 14, 3601–3605.
