To provide resiliency against pandemics, new types of universal biosensors that can be rapidly adapted and deployed on a large scale for the detection of new pathogens and new variants of known pathogens and exhibit real-time detection with LOD as low as a few target molecules need to be developed. Photonic biosensors, which provide optical signal amplification and can be mass-produced by existing semiconductor foundries, offer a promising platform, which is in principle amenable to multiplexing with spectroscopic and electrochemical detection techniques. However, to reach sub-fM detection levels, photonic biosensors typically have had to be engineered to support ultra-high-Q trapped modes.
- biosensors
- pathogen detection
- rapid sensing
- optical amplification
- point-of-care testing
1. Introduction
To achieve both selectivity and sensitivity of detection, bio(chemical) sensors typically require a target recognition element that has a strong affinity with the molecular target analyte in the tested sample, a transducer that generates a signal based on the presence of an analyte, which can be detected and processed, and an amplifier that increases the signal level, thus decreasing the minimum amount of target molecules that can be reliably detected. Biosensors with photonic transducers generate optical signals, which carry the information on either the presence or absence of the target analyte in the biological sample. These optical signals can be generated as a result of linear and/or non-linear light–matter interactions, including effective refractive index changes, increased light absorption, fluorescence, and Raman or Brillouin scattering. Small size, weak non-linear response, and low refractive index values of biological molecules of interest severely limit the intensity of any optical signal they can generate. Detection becomes even more challenging when only a few copies of these molecules are present in the tested sample, which is often the case in the early disease diagnostic efforts.
To address these challenges, various transducers and signal amplification schemes have been developed and optimized in the past decades. Despite many specific device realizations, descriptions of which can be found in recent review articles [1][2][3][4][5], the unique mechanisms of the observable optical signal generation include: (i) local changes to the light wave propagation and dissipation caused by the capture of the target molecules ( Figure 1 a), (ii) native non-linear signatures of target molecules revealed by optical excitation ( Figure 1 b), and (iii) target-activated release of optical labels that are the actual sources of the observable optical signals ( Figure 1 c). The affinity-type sensors shown in Figure 1 a evaluate selective target capture events and, while they can be used in the label-free format, require development of target-specific receptors to achieve selectivity [5][6][7]. The detection of newly-emerged or mutated viral strains requires the development of new receptors to maintain specificity.
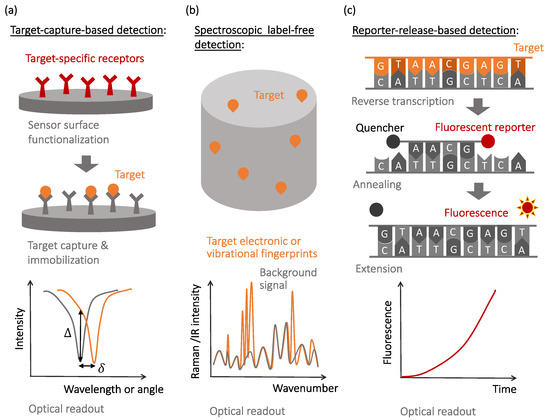
In contrast, spectroscopic label-free detection techniques, schematically illustrated in Figure 1 b, enable reading the native optical signature of the target molecules, which do not necessarily need to be selectively captured by the sensor surfaces [8]. However, nonlinear light–matter interactions that cause the generation of native optical fingerprints of target biomolecules are weak and often require optical or plasmonic amplification [9][10][11][12].
Finally, molecular recognition events that yield selectivity to the reporter-release biosensors are often similar to those used in the affinity-type sensors, with the observable signals being produced when optically-bright reporters are released during the recognition process and start generating optical signals (most often, fluorescence or visible color changes). This mechanism is illustrated in Figure 1 c with a specific example of the reverse transcription polymerase chain reaction (RT-PCR), currently considered the gold standard for SARS-CoV-2 detection [13][14][15].
2. Affinity-Type Biosensors with Optical Signal Amplification
The simplest interference-type photonic biosensor is a planar vertically interrogated Fabry–Perot interferometer ( Figure 2 a), which creates an interference pattern by combining the phase-shifted light reflected from different surfaces within the structure [16][17]. The spectral shifts and the periodicity changes of the interferometric pattern caused by the target molecules adsorption are evaluated during the detection process. Materials permeable to the target molecules (e.g., porous silicon or polymers) allow for a larger optical phase accumulation and thus exhibit more pronounced optical spectral shifts, but only if the concentration of target molecules or the volume of the analyte is large enough (or both), allowing for the capture of many molecules. This is not a situation observed in the early decrease diagnostics efforts, where only limited-volume samples containing femtomolar (fM) or sub-fM concentrations of target molecules are available for the analysis.
However, since molecule polarization depends not only on its polarizability but also on the strength of the excitation electric field, optical amplification techniques can improve the detection sensitivity. The optical amplification step is typically achieved in photonic biosensors via the transducer’s optical design [18]. The local electric field intensity can be increased by engineering the photonic transducer to trap light, localizing it to a micro- or nano-scale volume, and recycling the optical energy in the form of either a volumetric resonant mode or a surface polariton mode. Light trapping and guiding in high-contrast dielectric or metal structures enables both sensing volume miniaturization and optical amplification [19][20][21]. Guided-mode sensors can be realized with structures, such as metal–dielectric interfaces, supporting surface plasmon polariton modes or integrated Si waveguides. These sensing structures can be implemented in prism configurations or in waveguides, such as optical fibers ( Figure 2 c). Optical fiber-based sensors have been implemented for a variety of applications, including antibody [22], bacteria [23], and environmental sensing [24][25]. Fiber optic sensors offer many potential advantages over conventional planar structures, including their simplicity, low cost, and potential for multiplexing [26]. However, additional optical signal amplification is often required, which can be achieved via fiber-integrated structures supporting surface plasmon modes.
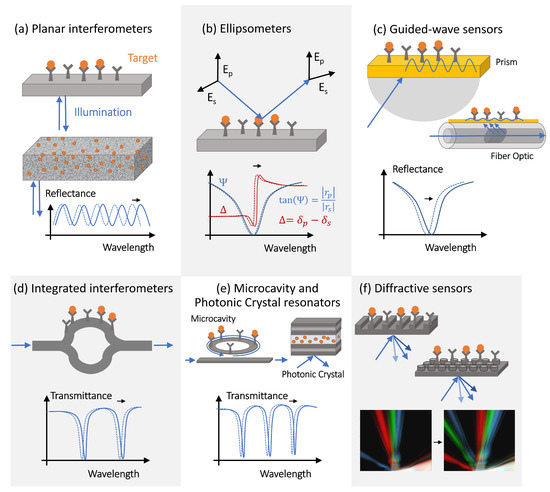
PhC cavities and other optical microcavity structures exhibit sharp resonant features in their transmission spectra as a result of the light interference between the reference signal propagating through the bus waveguide and the light trapped in the resonator optical mode [28]. These resonant spectral features are characterized by high-quality (Q) factors, defined as the ratio of the resonant frequency to resonance bandwidth [29][30], and shift in response to the resonator surface chemistry changes [31][28], providing a sensitive optical transduction mechanism.
Finally, all the label-free biosensors with optical signal amplification require chemical surface functionalization to immobilize target-specific receptors [18] and are vulnerable to non-specific binding [32][33][34]. This greatly decreases the sensor performance and reliability, as non-specific binding increases background signals and the sensor limit of detection. Although self-referencing techniques can somewhat improve the sensor performance by reducing the effect of the non-specific binding [35][36][37][38], incorporating self-referencing schemes in the sensor design increases the sensor cost and complexity. Another way to reduce the effect of non-specific binding and the LOD is by increasing the signal-to-noise ratio via adding labels that selectively bind to the target molecules, thus increasing the optical signal associated with the target capture event. Just like the surface receptors for the molecular target capture, these labels need to be custom-engineered to provide target selectivity and added to the assay together with the sample [39][40][41][42][43][44]. This further complicates the process and increases the initial development time of the sensor technology. Generally, SPR and LSPR can show real-time detection [45]; however, adding labels to reduce the LOD will increase the detection time because more time will be needed for particle diffusion [46].
3. Label-Free Spectroscopic Sensors with Digital and Optical Amplification
Some of the limitations of affinity-type refractometric optical biosensors can be overcome using vibrational spectroscopic techniques, such as infrared (IR) [47][48] and Raman spectroscopy ( Figure 3 ) [10][49]. These techniques measure the nonlinear interactions of the atoms and molecules present in the sample with incident electromagnetic radiation. The outcome is a spectral ’fingerprint’ of the sample that contains the information about its content, which is, most often, a vibrational spectrum of the biological molecule. While these direct ’fingerprinting’ methods can be combined with the use of IR-active or Raman-active labels, they typically are used to directly detect the native biological targets present in the sample [50][51]. This not only eliminates the need for multiple complex reagents, but also the physics involved in these processes are inherently faster than the ones requiring electrochemical reactions, and the sample preparation can be minimal. Furthermore, no prior knowledge of the composition of the sample is required in principle, as the target recognition step is performed digitally, by analyzing the acquired vibrational spectra. Therefore, vibration spectroscopy has the potential for fast, non-invasive, multiplexed, and reagent- and label-free detection.
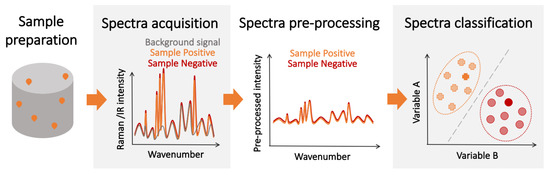
Regarding the SNR, its value is completely dependent on the experimental setup, and a large variety of spectrometers exist with different levels of SNR, making the determination of an overall “technique limit of detection” particularly challenging. However, some studies have been done to estimate and test these limits for certain analytes and under specific conditions [52][53][54]. However, since the spectrum used for detection contains information about all the IR/Raman-active biological components in the sample, imbalances in the content of the sample caused by disease-induced metabolic changes can be detected, enhancing the overall detection performance. Nevertheless, while the individual spectra of IR absorption and Raman scattering show greater variation between different biological species than their corresponding optical spectra, they may be hard to resolve since some of the vibrational bands might be common to other compounds of the sample. Thus, the ultimate performance of vibration spectroscopy-based detectors is not only determined by the limit of detection for the molecules of the pathogen. Given the aforementioned intricacies, experimental determination remains the best approach to assess the potential of these techniques for diagnosis.
While the sensitivity and specificity of spectroscopic detection methods can be increased by using the same target-specific receptors as those used in label-free affinity-type sensors, this would increase the sensor complexity, limit its use as a multiple pathogen detection platform, and may introduce additional noise from receptor-generated spectroscopic signatures. A potential alternative is to implement a two-step detection scheme where patients are first screened using fast spectroscopic techniques to identify likely positive candidates . This preliminary diagnosis could then be confirmed by a second more sensitive, slower, and resource-intensive detection method (e.g., PCR). This scheme could increase the overall number of tests and optimize the allocation of resources, which is essential during pandemics and in regions with limited resources. The advantage of less-sensitive, mass-deployable detection techniques for pandemic management has been explored in recent years [55][56]. Furthermore, chemometric techniques are often leveraged to facilitate the spectral analysis and classification [57]. Over the last several years, the combination of vibrational spectroscopy and chemometrics has been successfully demonstrated for the detection of various pathogens [58][59]. Spectroscopic techniques could also be used in combination with biological amplification for increased performance at the expense of detection time.
Finally, optical and digital amplification techniques have been successfully implemented to enhance sensitivity and decrease the LOD of spectroscopic detection techniques. For example, several stimulated Raman techniques, such as coherent anti-Stokes Raman scattering [60] or stimulated Raman scattering [61], have been developed, and the potential of Raman spectroscopy for pathogen detection has been studied and demonstrated in recent decades [62][59][63][64]. Similar to the case of affinity-type biosensors, both the Raman signal and the infrared absorption efficiency can be enhanced by the excitation of the surface plasmon polariton modes on the sensing surfaces. Since the signal scales with the product of the optical field intensities at the excitation and the Raman scattering wavelengths, plasmon-enhanced local field amplification can yield orders-of-magnitude signal enhancement. In particular, two techniques have emerged as particularly promising for bioanalysis, which are surface-enhanced Raman spectroscopy (SERS) [65][66][67][68] and tip-enhanced Raman spectroscopy (TERS) [69]. These techniques have already been proven effective for pathogen detection [64][70][71]. Overall, one of the greatest advantages of vibrational spectroscopy is its potential use for the multiplexed detection of a variety of pathogens, in combination with affinity-type optical detection techniques.
4. Conclusions and Outlook
To provide resiliency against pandemics, new types of universal biosensors that can be rapidly adapted and deployed on a large scale for the detection of new pathogens and new variants of known pathogens and exhibit real-time detection with LOD as low as a few target molecules need to be developed. Photonic biosensors, which provide optical signal amplification and can be mass-produced by existing semiconductor foundries, offer a promising platform, which is in principle amenable to multiplexing with spectroscopic and electrochemical detection techniques. However, to reach sub-fM detection levels, photonic biosensors typically have had to be engineered to support ultra-high-Q trapped modes. On the other hand, the plasmonic field enhancement required for SERS, infrared and fluorescent signal enhancement is accompanied by the inevitable Q-factor reduction due to Ohmic losses in metals, hindering multiplexing opportunities. Hybrid optoplasmonic biosensors offer a partial solution to this dilemma [72][73][74][75][76] but have not been adopted on larger scale owing to their complexity in fabrication and operation. Finally, even ’label-free’ photonic sensors typically rely on target-specific receptors to achieve specificity and increase sensitivity and need to be re-designed for detection of new pathogens.
The recently introduced concept of the probe-cleavage detection makes the rapid development of new highly sensitive real-time photonic biosensor platforms possible for emergent pathogens and new variants of known pathogens because it no longer requires the development of target-specific receptors and offers low detection limits (down to few molecules) owing to the high collateral cleavage counts exhibited by the commercially available programmable CRISPR– Cas complexes [77][78][79][80][81].
Interestingly, both low- and high-quality-factor photonic biosensors can be re-engineered to work efficiently under the probe-cleavage detection scenario owing to the dramatic signal amplification provided by the high-contrast probes and collateral cleavage multiplication factors [82][83][84]. This offers opportunities for multiplexing photonic biosensors with spectroscopic techniques and making use of the surface-plasmon signal enhancement. Furthermore, low- and moderate-Q photonic biosensor platforms can be engineered to operate with the single-frequency fixed-angle intensity vertical readout, reducing their cost and complexity and enabling integration with portable smartphone-based systems [85][86][87]. We expect that many previously developed photonic biosensing platforms can be redesigned to operate in the probe-cleavage detection regime and provide multiplexed capabilities. The recently developed toolbox of the inverse design and machine-learning techniques [84][88][89][90][91][92] can be utilized to find the optimum sensor configuration (i.e., a photonic device platform, readout mechanism, type and surface coverage of the nanoparticle probes, etc.) that balances the sensor ease of use, detection time, LOD, and the dynamic range requirements.
This entry is adapted from the peer-reviewed paper 10.3390/photonics8080342
References
- González-Guerrero, A.; Dante, S.; Duval, D.; Osmond, J.; Lechuga, L.M. Advanced photonic biosensors for point-of-care diagnostics. Procedia Eng. 2011, 25, 71–75.
- Luan, E.; Shoman, H.; Ratner, D.M.; Cheung, K.C.; Chrostowski, L. Silicon photonic biosensors using label-free detection. Sensors 2018, 18, 3519.
- Vollmer, F.; Yang, L. Review Label-free detection with high-Q microcavities: A review of biosensing mechanisms for integrated devices. Nanophotonics 2012, 1, 267–291.
- Chen, C.; Wang, J. Optical biosensors: An exhaustive and comprehensive review. Analyst 2020, 145, 1605–1628.
- Huertas, C.S.; Calvo-Lozano, O.; Mitchell, A.; Lechuga, L.M. Advanced evanescent-wave optical biosensors for the detection of nucleic acids: An analytic perspective. Front. Chem. 2019, 7, 724.
- Nangare, S.N.; Patil, P.O. Affinity-Based Nanoarchitectured Biotransducer for Sensitivity Enhancement of Surface Plasmon Resonance Sensors for In Vitro Diagnosis: A Review. ACS Biomater. Sci. Eng. 2020, 7, 2–30.
- Soler, M.; Estevez, M.C.; Cardenosa-Rubio, M.; Astua, A.; Lechuga, L.M. How nanophotonic label-free biosensors can contribute to rapid and massive diagnostics of respiratory virus infections: COVID-19 case. ACS Sens. 2020, 5, 2663–2678.
- Cheng, Y.; Dong, L.; Zhang, J.; Zhao, Y.; Li, Z. Recent advances in microRNA detection. Analyst 2018, 143, 1758–1774.
- Langer, J.; Jimenez de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguié, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.; Boisen, A.; Brolo, A.G.; et al. Present and future of surface-enhanced Raman scattering. ACS Nano 2019, 14, 28–117.
- Pilot, R.; Signorini, R.; Durante, C.; Orian, L.; Bhamidipati, M.; Fabris, L. A review on surface-enhanced Raman scattering. Biosensors 2019, 9, 57.
- Stockman, M.I.; Kneipp, K.; Bozhevolnyi, S.I.; Saha, S.; Dutta, A.; Ndukaife, J.; Kinsey, N.; Reddy, H.; Guler, U.; Shalaev, V.M.; et al. Roadmap on plasmonics. J. Opt. 2018, 20, 043001.
- Zhou, C.; Zou, H.; Sun, C.; Ren, D.; Chen, J.; Li, Y. Signal amplification strategies for DNA-based surface plasmon resonance biosensors. Biosens. Bioelectron. 2018, 117, 678–689.
- Zhao, S. Alternative splicing, RNA-seq and drug discovery. Drug Discov. Today 2019, 24, 1258–1267.
- Tahamtan, A.; Ardebili, A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Rev. Mol. Diagn. 2020, 20, 453–454.
- Van Kasteren, P.B.; van Der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.B.; Meijer, A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020, 128, 104412.
- Lin, V.S. A Porous Silicon-Based Optical Interferometric Biosensor. Science 1997, 278, 840–843.
- Arshavsky-Graham, S.; Massad-Ivanir, N.; Segal, E.; Weiss, S. Porous Silicon-Based Photonic Biosensors: Current Status and Emerging Applications. Anal. Chem. 2019, 91, 441–467.
- Chen, Y.; Liu, J.; Yang, Z.; Wilkinson, J.S.; Zhou, X. Optical biosensors based on refractometric sensing schemes: A review. Biosens. Bioelectron. 2019, 144, 111693.
- Wu, L.; Guo, J.; Xu, H.; Dai, X.; Xiang, Y. Ultrasensitive biosensors based on long-range surface plasmon polariton and dielectric waveguide modes. Photonics Res. 2016, 4, 262.
- Fan, B.; Liu, F.; Li, Y.; Wang, X.; Cui, K.; Feng, X.; Zhang, W.; Huang, Y. Integrated refractive index sensor based on hybrid coupler with short range surface plasmon polariton and dielectric waveguide. Sens. Actuators B Chem. 2013, 186, 495–505.
- Piliarik, M.; Vaisocherová, H.; Homola, J. Surface plasmon resonance biosensing. Biosens. Biodetect. 2009, 65–88.
- Zeni, L.; Perri, C.; Cennamo, N.; Arcadio, F.; D’Agostino, G.; Salmona, M.; Beeg, M.; Gobbi, M. A portable optical-fibre-based surface plasmon resonance biosensor for the detection of therapeutic antibodies in human serum. Sci. Rep. 2020, 10, 11154.
- Kumar, S.; Guo, Z.; Singh, R.; Wang, Q.; Zhang, B.; Cheng, S.; Liu, F.Z.; Marques, C.; Kaushik, B.K.; Jha, R. MoS2 Functionalized Multicore Fiber Probes for Selective Detection of Shigella Bacteria Based on Localized Plasmon. J. Light. Technol. 2021, 39, 4069–4081.
- Cai, S.; Pan, H.; González-Vila, Á.; Guo, T.; Gillan, D.C.; Wattiez, R.; Caucheteur, C. Selective detection of cadmium ions using plasmonic optical fiber gratings functionalized with bacteria. Opt. Express 2020, 28, 19740.
- Wang, Y.; Zhu, G.; Li, M.; Singh, R.; Marques, C.; Min, R.; Kaushik, B.K.; Zhang, B.; Jha, R.; Kumar, S. Water pollutants p-Cresol detection based on Au-ZnO nanoparticles modified tapered optical fiber. IEEE Trans. Nanobiosci. 2021, 20, 377–384.
- Zhao, Y.; Tong, R.j.; Xia, F.; Peng, Y. Current status of optical fiber biosensor based on surface plasmon resonance. Biosens. Bioelectron. 2019, 142, 111505.
- El-Aziz, O.A.; Abd Elsayed, H.A.; Sayed, M. One-dimensional defective photonic crystals for the sensing and detection of protein. Appl. Opt. 2019, 58, 8309–8315.
- Su, J. Label-Free Biological and Chemical Sensing Using Whispering Gallery Mode Optical Resonators: Past, Present, and Future. Sensors 2017, 17, 540.
- Paschotta, R. Q factor. In Encyclopedia of Laser Physics and Technology, 1st ed.; Wiley-VCH: Weinheim, Germany, 2008.
- Braginsky, V.; Gorodetsky, M.; Ilchenko, V. Quality-factor and nonlinear properties of optical whispering-gallery modes. Phys. Lett. A 1989, 137, 393–397.
- Wang, Y.; Zeng, S.; Humbert, G.; Ho, H. Microfluidic Whispering Gallery Mode Optical Sensors for Biological Applications. Laser Photonics Rev. 2020, 14, 2000135.
- Lichtenberg, J.Y.; Ling, Y.; Kim, S. Non-Specific Adsorption Reduction Methods in Biosensing. Sensors 2019, 19, 2488.
- Johnson, B.N.; Mutharasan, R. Biosensor-based microRNA detection: Techniques, design, performance, and challenges. Analyst 2014, 139, 1576–1588.
- Liu, B.; Liu, X.; Shi, S.; Huang, R.; Su, R.; Qi, W.; He, Z. Design and mechanisms of antifouling materials for surface plasmon resonance sensors. Acta Biomater. 2016, 40, 100–118.
- Zourob, M.; Elwary, S.; Turner, A.; Campbell, D.P. Interferometric Biosensors. In Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems; Springer: Berlin/Heidelberg, Germany, 2008.
- Goodwin, M.J.; Besselink, G.A.J.; Falke, F.; Everhardt, A.S.; Cornelissen, J.J.L.M.; Huskens, J. Highly Sensitive Protein Detection by Asymmetric Mach–Zehnder Interferometry for Biosensing Applications. ACS Appl. Bio Mater. 2020, 3, 4566–4572.
- Li, M.; Wu, X.; Liu, L.; Fan, X.; Xu, L. Self-Referencing Optofluidic Ring Resonator Sensor for Highly Sensitive Biomolecular Detection. Anal. Chem. 2013, 85, 9328–9332.
- Boriskina, S.V.; Negro, L.D. Self-referenced photonic molecule bio(chemical)sensor. Opt. Lett. 2010, 35, 2496–2498.
- Jang, H.S.; Park, K.N.; Kang, C.D.; Kim, J.P.; Sim, S.J.; Lee, K.S. Optical fiber SPR biosensor with sandwich assay for the detection of prostate specific antigen. Opt. Commun. 2009, 282, 2827–2830.
- Seo, H.B.; Gu, M.B. Aptamer-based sandwich-type biosensors. J. Biol. Eng. 2017, 11, 1–7.
- Seok, J.S.; Ju, H. Plasmonic Optical Biosensors for Detecting C-Reactive Protein: A Review. Micromachines 2020, 11, 895.
- Wu, B.; Jiang, R.; Wang, Q.; Huang, J.; Yang, X.; Wang, K.; Li, W.; Chen, N.; Li, Q. Detection of C-reactive protein using nanoparticle-enhanced surface plasmon resonance using an aptamer-antibody sandwich assay. Chem. Commun. 2016, 52, 3568–3571.
- Meyer, M.H.; Hartmann, M.; Keusgen, M. SPR-based immunosensor for the CRP detection—A new method to detect a well known protein. Biosens. Bioelectron. 2006, 21, 1987–1990.
- Vance, S.A.; Sandros, M.G. Zeptomole Detection of C-Reactive Protein in Serum by a Nanoparticle Amplified Surface Plasmon Resonance Imaging Aptasensor. Sci. Rep. 2015, 4, 5129.
- Singh, P. SPR Biosensors: Historical Perspectives and Current Challenges. Sens. Actuators B Chem. 2016, 229, 110–130.
- Sheehan, P.E.; Whitman, L.J. Detection Limits for Nanoscale Biosensors. Nano Lett. 2005, 5, 803–807.
- Beć, K.B.; Grabska, J.; Huck, C.W. Biomolecular and bioanalytical applications of infrared spectroscopy—A review. Anal. Chim. Acta 2020, 1133, 150–177.
- Ewing, G.E. Infrared Spectroscopy. Annu. Rev. Phys. Chem. 1972, 23, 141–164.
- Notingher, I. Raman Spectroscopy Cell-based Biosensors. Sensors 2007, 7, 1343.
- Chen, Y.; Qian, C.; Liu, C.; Shen, H.; Wang, Z.; Ping, J.; Wu, J.; Chen, H. Nucleic acid amplification free biosensors for pathogen detection. Biosens. Bioelectron. 2020, 153, 112049.
- Balan, V.; Mihai, C.T.; Cojocaru, F.D.; Uritu, C.M.; Dodi, G.; Botezat, D.; Gardikiotis, I. Vibrational spectroscopy fingerprinting in medicine: From molecular to clinical practice. Materials 2019, 12, 2884.
- Khoshmanesh, A.; Dixon, M.W.; Kenny, S.; Tilley, L.; McNaughton, D.; Wood, B.R. Detection and quantification of early-stage malaria parasites in laboratory infected erythrocytes by attenuated total reflectance infrared spectroscopy and multivariate analysis. Anal. Chem. 2014, 86, 4379–4386.
- Theakstone, A.G.; Rinaldi, C.; Butler, H.J.; Cameron, J.M.; Confield, L.R.; Rutherford, S.H.; Sala, A.; Sangamnerkar, S.; Baker, M.J. Fourier-transform infrared spectroscopy of biofluids: A practical approach. Transl. Biophotonics 2021, 3, e202000025.
- Parachalil, D.R.; Bruno, C.; Bonnier, F.; Blasco, H.; Chourpa, I.; Baker, M.J.; McIntyre, J.; Byrne, H.J. Analysis of bodily fluids using vibrational spectroscopy: A direct comparison of Raman scattering and infrared absorption techniques for the case of glucose in blood serum. Analyst 2019, 144, 3334–3346.
- Mina, M.J.; Parker, R.; Larremore, D.B. Rethinking Covid-19 test sensitivity—A strategy for containment. N. Engl. J. Med. 2020, 383, e120.
- Mina, M.J.; Andersen, K.G. COVID-19 testing: One size does not fit all. Science 2021, 371, 126–127.
- Morais, C.L.; Lima, K.M.; Singh, M.; Martin, F.L. Tutorial: Multivariate classification for vibrational spectroscopy in biological samples. Nat. Protoc. 2020, 15, 2143–2162.
- Santos, M.C.; Morais, C.L.; Nascimento, Y.M.; Araujo, J.M.; Lima, K.M. Spectroscopy with computational analysis in virological studies: A decade (2006–2016). TrAC Trends Anal. Chem. 2017, 97, 244–256.
- Ralbovsky, N.M.; Lednev, I.K. Towards development of a novel universal medical diagnostic method: Raman spectroscopy and machine learning. Chem. Soc. Rev. 2020, 49, 7428–7453.
- Evans, C.L.; Xie, X.S. Coherent anti-Stokes Raman scattering microscopy: Chemical imaging for biology and medicine. Annu. Rev. Anal. Chem. 2008, 1, 883–909.
- Freudiger, C.W.; Min, W.; Saar, B.G.; Lu, S.; Holtom, G.R.; He, C.; Tsai, J.C.; Kang, J.X.; Xie, X.S. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 2008, 322, 1857–1861.
- Kong, K.; Kendall, C.; Stone, N.; Notingher, I. Raman spectroscopy for medical diagnostics—From in-vitro biofluid assays to in-vivo cancer detection. Adv. Drug Deliv. Rev. 2015, 89, 121–134.
- Lambert, P.J.; Whitman, A.G.; Dyson, O.F.; Akula, S.M. Raman spectroscopy: The gateway into tomorrow’s virology. Virol. J. 2006, 3, 51.
- Austin, L.A.; Osseiran, S.; Evans, C.L. Raman technologies in cancer diagnostics. Analyst 2016, 141, 476–503.
- Paraskevaidi, M.; Ashton, K.M.; Stringfellow, H.F.; Wood, N.J.; Keating, P.J.; Rowbottom, A.W.; Martin-Hirsch, P.L.; Martin, F.L. Raman spectroscopic techniques to detect ovarian cancer biomarkers in blood plasma. Talanta 2018, 189, 281–288.
- Moore, T.J.; Moody, A.S.; Payne, T.D.; Sarabia, G.M.; Daniel, A.R.; Sharma, B. In vitro and in vivo SERS biosensing for disease diagnosis. Biosensors 2018, 8, 46.
- Zong, C.; Xu, M.; Xu, L.J.; Wei, T.; Ma, X.; Zheng, X.S.; Hu, R.; Ren, B. Surface-enhanced Raman spectroscopy for bioanalysis: Reliability and challenges. Chem. Rev. 2018, 118, 4946–4980.
- Henry, A.I.; Sharma, B.; Cardinal, M.F.; Kurouski, D.; Duyne, R.P.V. Surface-Enhanced Raman Spectroscopy Biosensing: Vivo Diagn. Multimodal Imaging. Anal. Chem. 2016, 88, 6638–6647.
- Hermann, P.; Hermelink, A.; Lausch, V.; Holland, G.; Möller, L.; Bannert, N.; Naumann, D. Evaluation of tip-enhanced Raman spectroscopy for characterizing different virus strains. Analyst 2011, 136, 1148–1152.
- Lim, J.Y.; Nam, J.S.; Shin, H.; Park, J.; Song, H.I.; Kang, M.; Lim, K.I.; Choi, Y. Identification of newly emerging influenza viruses by detecting the virally infected cells based on surface enhanced Raman spectroscopy and principal component analysis. Anal. Chem. 2019, 91, 5677–5684.
- Cialla, D.; Deckert-Gaudig, T.; Budich, C.; Laue, M.; Möller, R.; Naumann, D.; Deckert, V.; Popp, J. Raman to the limit: Tip-enhanced Raman spectroscopic investigations of a single tobacco mosaic virus. J. Raman Spectrosc. Int. J. Orig. Work All Asp. Raman Spectrosc. Incl. High. Order Process. Also Brillouin Rayleigh Scatt. 2009, 40, 240–243.
- Santiago-Cordoba, M.A.; Boriskina, S.V.; Vollmer, F.; Demirel, M.C. Nanoparticle-based protein detection by optical shift of a resonant microcavity. Appl. Phys. Lett. 2011, 99, 073701.
- Santiago-Cordoba, M.A.; Cetinkaya, M.; Boriskina, S.V.; Vollmer, F.; Demirel, M.C. Ultrasensitive detection of a protein by optical trapping in a photonic-plasmonic microcavity. J. Biophotonics 2012, 5, 629–638.
- Shopova, S.I.; Rajmangal, R.; Holler, S.; Arnold, S. Plasmonic enhancement of a whispering-gallery-mode biosensor for single nanoparticle detection. Appl. Phys. Lett. 2011, 98, 243104.
- Hong, Y.; Reinhard, B.M. Optoplasmonics: Basic principles and applications. J. Opt. 2019, 21, 113001.
- Baaske, M.D.; Foreman, M.R.; Vollmer, F. Single-molecule nucleic acid interactions monitored on a label-free microcavity biosensor platform. Nat. Nanotechnol. 2014, 9, 933–939.
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR—Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874.
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 1–10.
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Simon, H.Y.; Boehm, C.K.; Kosoko-Thoroddsen, T.S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282.
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012.
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149.
- Layouni, R.; Dubrovsky, M.; Bao, M.; Chung, H.; Du, K.; Boriskina, S.V.; Weiss, S.M.; Vermeulen, D. High contrast cleavage detection for enhancing porous silicon sensor sensitivity. Opt. Express 2021, 29, 1.
- Dubrovsky, M.; Blevins, M.; Boriskina, S.V.; Vermeulen, D. High Contrast Cleavage Detection. Opt. Lett. 2021, 46, 2593–2596.
- Chung, H.; Boriskina, S.V. Inverse design of a single-frequency diffractive biosensor based on the reporter cleavage detection mechanism. Opt. Express 2021, 29, 10780.
- Blevins, M.G.; Michel, A.P.M.; Boriskina, S.V. Planar nanophotonic structures for intensity based readout refractive index sensing applied to dissolved methane detection. OSA Contin. 2020, 3, 3556.
- Nazirizadeh, Y.; Bog, U.; Sekula, S.; Mappes, T.; Lemmer, U.; Gerken, M. Low-cost label-free biosensors using photonic crystals embedded between crossed polarizers. Opt. Express 2010, 18, 19120.
- Lin, Y.C.; Hsieh, W.H.; Chau, L.K.; Chang, G.E. Intensity-detection-based guided-mode-resonance optofluidic biosensing system for rapid, low-cost, label-free detection. Sens. Actuators B Chem. 2017, 250, 659–666.
- Kim, D.C.; Hermerschmidt, A.; Dyachenko, P.; Scharf, T. Inverse design and demonstration of high-performance wide-angle diffractive optical elements. Opt. Express 2020, 28, 22321.
- Liu, Z.; Zhu, D.; Raju, L.; Cai, W. Tackling Photonic Inverse Design with Machine Learning. Adv. Sci. 2021, 8, 2002923.
- Chung, H.; Miller, O.D. High-NA achromatic metalenses by inverse design. Opt. Express 2019, 28, 6945–6965.
- Colburn, S.; Majumdar, A. Inverse design and flexible parameterization of meta-optics using algorithmic differentiation. Commun. Phys. 2021, 4, 1–11.
- Ma, W.; Liu, Z.; Kudyshev, Z.A.; Boltasseva, A.; Cai, W.; Liu, Y. Deep learning for the design of photonic structures. Nat. Photonics 2021, 15, 77–90.
