Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Engineering, Biomedical
Wastewater treatment plants and other remediation facilities serve important roles, both in public health, but also as dynamic research platforms for acquiring useful resources and biomolecules for various applications. An example of this is methanotrophic bacteria within anaerobic digestion processes in wastewater treatment plants. These bacteria are an important microbial source of many products including ectoine, polyhydroxyalkanoates, and methanobactins, which are invaluable to the fields of biotechnology and biomedicine.
- methanotrophs
- methanobactin
- ectoine
- biogas
- S-layer
- polyhydroxyalkonate (PHA)
- polyhydroxybutyrate (PHB)
- methane monooxygenase (MMO)
- exopolysaccharide (EPS)
- single cell protein (SCP)
- wastewater treatment
- biomedical applications
1. Introduction
In recent years, a global movement has engaged targeting the development of alternative bio-based therapeutic products for biomedical applications in order to reduce or eliminate the adverse side effects associated with the use of non-biocompatible compounds by the human immune system [1][2]. A broad spectrum of naturally occurring compounds derived from animals, plants, or microbes has been tested for their employment in modern medicine [3]. In the early 2000s, twenty naturally derived therapeutic drugs were developed and brought to market [4], though this clearly was not the first instance of using biologically-derived molecules in medicine. The use of naturally occurring compounds has been documented in ancient civilizations such as Egypt and China, where they relied on plant extracts and honey for remediation and healing purposes [5], and of course Alexander Fleming’s discovery of penicillin opened the door for the use of antibiotics to fight infection, but also highlighted the potential for microbially synthesized products in the pharmaceutical and medical industries [6]. The employment of bioactive composites for use in many industries has become a more robust and efficient process in many applications including: (i) the manufacturing of biopolymers, nanoparticles, pigments for the productions of drug capsules, optical fibers, electronic devices, and paint [7][8][9], (ii) the extraction of catalytic enzymes, organic acids, and surfactants to employ in drug, food, and soap industries [10][11][12], and (iii) the use of organic vitamins, lipids, and proteins, and microbial metabolites to generate synthetic hormones, nutritional supplements, targeted therapies, vaccines anti-cancer agents, anti-inflammatory drugs, and the overall medicinal industry [13][14][15][16][17]. Interestingly, wastewater treatment plants (WWTPs) comprise an important asset for its public health, environmental and economic contributions, in addition to their function as a remediation facility. WWTPs encompass an important revenue stream for their role in resource recovery [18]. More recently, WWTPs have been viewed as biorefinery facilities; they exploit the organic matter within wastewater as microbial substrates in order to sustainably generate electricity, remove contaminants, and recover resources [19]. WWTPs can be considered large biodiverse microbial populations that are distinct within each stage of the water treatment process. Each microbial population can also be characterized with the ability to produce a variety of value-added products, each of which is suitable for use in a variety of implementations in different sectors of biomedical industries [18][20]. WWTPs have a broad variety of different important bacterial genera such as purple sulfur bacteria, ammonia oxidizing bacteria, nitrate oxidizing bacteria, Pseudomonas, Mycobacterium, and Methylobacterium [21]. Each bacterial population represents important contributors for resource recovery for the production of biopolymers, catalytic enzymes, lipids, and proteins [19]. While bioproduct resource recovery from WWTPs can be more challenging compared to purely synthetic methods, it is a more sustainable option and is essential to overcome limitations in resource availability. For instance, more synthetic routes for production of high-value biomolecules require cost-intensive processes and bio-refineries for realistic applications. On the other hand, while there are many technical challenges in the processes of optimal bacterial cultivation, biomolecule extraction, and purification from resource pools such as WWTPs, many of these challenges are offset by the large and renewable feedstock, leading to lower input costs associated with bioproduct development. In this review we highlight the role of methane oxidizing bacteria, namely the methanotrophs found in anerobic digestion processes of WWTPs, as a multiple high-value bioproduct generating system with diverse potential biomedical applications. Furthermore, we provide an overview of the enzymatic pathways employed by methanotrophs to generate different metabolites and demonstrate the dynamic interactions of different types of biomolecules.
2. Methanotrophic Bacteria in Water Treatment Systems
Methanotrophic bacteria have a unique metabolism that relies on a single carbon substrate, methane. However, its sophistication allows it to manufacture a mixture of value-added organic compounds. Methanotrophs utilize methane gas as an electron donor in order to produce sufficient energy required for cellular growth and biosynthesis of different metabolites [22]. While methanotrophs might not be a top producer for certain products, this is compensated by its ability to perform multiple roles simultaneously which begins with the oxidation of methane gas to produce methanol, an important biofuel [23]. Methane gas is the second most potent greenhouse gas after carbon dioxide; therefore, methanotrophic bacteria’s ability to mitigate this harmful biogas is crucial in the global methane cycle. WWTPs are responsible for an estimated 4% of the global methane production; the gas is normally flared into the atmosphere contributing to global warming [24]. Whenever methanotrophs are present in WWTPs, they are associated with the availability of methane gas produced from the anaerobic digestion process, and with the right allocations the generated biogas can be exploited for resource recovery purposes [25].
Methanotrophs are Gram-negative bacteria and are a subgroup of a broader bacterial group known as methylotrophs [26]. They are distinct in their reliance on methane oxidization, unlike methylotrophs that have the potential to utilize different single carbon substrate such as methanol, halomethanes, and methylated amines [27]. Thus, aerobic methanotrophs metabolism relies on the role of oxygen to oxidize the methane substrate to ultimately generate carbon dioxide (CO2) and water [28].
There are three types of aerobic methanotrophs that differ phylogenetically, types I, II, or X, each of which follow a distinct metabolic pathway. All three types share a common methane oxidation pathway to produce formaldehyde, after which, each group carries on in a different enzymatic pathway. Type I methanotrophs employ the ribulose monophosphate (RuMP), and type II methanotrophs utilize the serine pathway (Figure 1) [29]. Type X methanotrophs exhibit similarities with type I methanotrophs in that they use the RuMP pathway; however, it differs from type I as they also have low concentrations of the serine pathway enzyme ribulose–bisphosphate carboxylase [30].
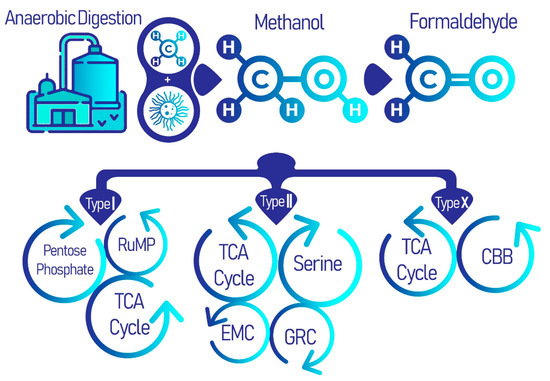
Figure 1. Methanotrophs in anaerobic digestion processes of WWTPs using the common methane oxidation pathway, which branches out for distinct metabolic cycles for each type of methanotrophs: types I, II, and X. It is these distinct secondary metabolic pathways, including the pentose-phosphate pathway (PPP), tricarboxylic acid (TCA) cycle, serine cycle, ethyl malonyl-CoA (EMC) pathway, glyoxylate regeneration cycle (GRC) and the Calvin-Benson-Bassham (CBB) cycle, that produce a diversity of useful biomolecules.
2.1. Taxonomy and Phenotype
Aerobic methanotrophs are taxonomically classified based on their phenotype, ability for spore formation, possession of specific membrane bound proteins, and their metabolic properties [24]. There are three main groups of methane oxidizing bacteria, types I, II, and X, each of which undertake a unique enzymatic pathway [31]. Type I and X methanotrophs belong to gamma-proteobacteria and reside in families Methylococcaceae and Methylothermaceae [32], whereas type II methanotrophs belong to alpha-proteobacteria from families Methylocystaceae and Beijerinckiaceae. Over time other groups of methanotrophs have emerged, including filamentous methane oxidizers with unusual methane monooxygenase, and extremely acidophilic bacteria of the phylum Verrucomicrobia. The classifications of methane-utilizing bacteria has of course evolved; however, the terms types I, II, and X are still commonly used when discussing the methanotrophs due to the distinctiveness of metabolic pathways for carbon fixation across phylogenetic groupings [26][30]. Morphologically, methanotrophs exhibit red coloring under Gram-staining and have various physical morphologies. For instance, type X methanotrophs are mainly found as paired cocci, while type II methanotrophs are crescent shaped rods and can occur in rosettes, and finally, type I methanotrophs can be found as either single cocci or rods [24].
2.2. EcoPhysiology
Aerobic methanotrophic bacteria inhabit oxic zones where oxygen is present as an electron acceptor and organic carbon, methane, is present for cellular biosynthesis, such as soils and freshwater, rice paddies, and WWTP sludge [33]. Most methanotrophs are mesophilic and prefer neutral pH; however, some thermophilic genera inhabit areas with high methane profusion and high temperatures such as volcanoes and soil paddies [34][35]. Moreover, a few psychrophilic species found in temperatures between 4 and 10 °C mainly belonging to type I methanotrophs have been reported in arctic regions [36][37][38].
Different types of methanotrophs follow distinct metabolic pathways after the oxidation of methane to formaldehyde and formate, which lead to the formation of industrially and biomedically valuable biomolecules and biopolymers as illustrated in Figure 2. First, methane is converted to methanol by the action of methane monooxygenase (MMO), then methanol dehydrogenase (MDH) further oxidizes methanol into formaldehyde [39]. There are two types of MMO: (i) the membrane bound and copper reliant particulate (p)MMO and (ii) soluble (s)MMO. Nonetheless, the pmoA gene encoding for pMMO is considered a universal marker for methanotrophic bacteria and is expressed by nearly all types of methanotrophs. Conversely, sMMO is typically expressed by type II and some type X methanotrophs under conditions of copper scarcity, where it utilizes iron as an alternative [40]. Following the common methane oxidation pathway, type I methanotrophs undergo the RuMP cycle, which is responsible for formaldehyde assimilation and detoxification. In the RuMP cycle, formaldehyde is fixed with ribulose 5-phsosphate to form 3-hexulose-6-phophate, which is then converted to glyceraldehyde-3-phosphate to finally generate pyruvate. There is an interplay between the RuMP pathway and the pentose phosphate pathway (PPP), where the latter ensures the regeneration of ribulose-5-phsosphate, while the former produces fructose-6-phosphote that is further metabolized via the PPP [41]. Thereafter, the pentose phosphate shunt is responsible for the generation of NADPH and ribose, which is of course important for the formation of nucleotide based biological molecules. These two pathways are crucial precursors of amino acid, nucleotide, and lipid biosynthesis. Pyruvate is further converted into acetyl-CoA for incorporation into the tricarboxylic acid (TCA) cycle and the electron transport chain for energy generation [27].
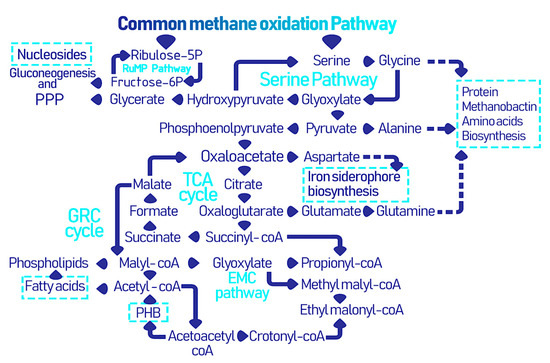
Figure 2. The collective metabolic pathways within methanotrophs leading to the formation of industrially and biomedically valuable biomolecules and biopolymers [25].
In type II methanotrophs, formaldehyde produced from the common metabolic pathway is further oxidized to formate and then converted to 5,10-methylenetetrahydrofolate to form serine and yields phosphoglycerate in several stepwise reactions of the serine cycle. The serine pathway requires 3 ATP and 2 NADH for activation unlike the RuMP cycle which only requires a single ATP. The serine pathway is simultaneously synced with the ethyl malonyl Co-A (EMC) pathway which includes several CoA thioesters, starting with malyl-CoA in the TCA which is converted into acetyl-CoA and eventually ethyl malonyl-CoA is cleaved into glyoxylate and propionyl-CoA where the former is an intermediate for the formation of oxaloacetate and succinyl-CoA for the latter, which are important originators for creating high-end products [42]. Furthermore, an intermediary cycle for glyoxylate recycling, the glyoxylate regeneration cycle (GRC), overlaps with the TCA and EMC cycles in type II methanotrophs and is considered as an additional route for glyoxylate regeneration [40][43]. Thus, the serine, EMC, and GRC pathways lead to mapping the derivatization of the secondary metabolites in type II methanotrophs. In nutrient-deficient conditions, an additional pathway to store energy in the form of polyhydroxybutyrate (PHB) granules is engaged [44].
This entry is adapted from the peer-reviewed paper 10.3390/biom11081217
References
- Kohane, D.S.; Langer, R. Polymeric biomaterials in tissue engineering. Pediatr. Res. 2008, 63, 487–491.
- Doltra, A.; Dietrich, T.; Schneeweis, C.; Kelle, S.; Doltra, A.; Stawowy, P.; Fleck, E. Magnetic Resonance Imaging of Cardiovascular Fibrosis and Inflammation: From Clinical Practice to Animal Studies and Back. BioMed Res. Int. 2013, 2013, 676489.
- Vaishnav, P.; Demain, A.L. Unexpected applications of secondary metabolites. Biotechnol. Adv. 2011, 29, 223–229.
- Chin, Y.-W.; Balunas, M.J.; Chai, H.B.; Kinghorn, A.D. Drug discovery from natural sources. AAPS J. 2006, 8, E239–E253.
- Subbarayappa, B.V. The roots of ancient medicine: An historical outline. J. Biosci. 2001, 26, 135–143.
- Singh, B.P.; Rateb, M.E.; Rodriguez-Couto, S.; Polizeli, M. de L.T. de M.; Li, W.-J. Editorial: Microbial Secondary Metabolites: Recent Developments and Technological Challenges. Front. Microbiol. 2019, 10, 914.
- Williams, S.F.; Martin, D.P. Applications of Polyhydroxyalkanoates (PHA) in Medicine and Pharmacy. Biopolym. Online 2005.
- Narsing Rao, M.P.; Xiao, M.; Li, W.J. Fungal and bacterial pigments: Secondary metabolites with wide applications. Front. Microbiol. 2017, 8, 1113.
- Baile, P.; Vidal, L.; Canals, A. A modified zeolite/iron oxide composite as a sorbent for magnetic dispersive solid-phase extraction for the preconcentration of nonsteroidal anti-inflammatory drugs in water and urine samples. J. Chromatogr. A 2019, 1603, 33–43.
- Ruiz, B.; Chávez, A.; Forero, A.; García-Huante, Y.; Romero, A.; Sánchez, M.; Rocha, D.; Sánchez, B.; Rodríguez-Sanoja, R.; Sánchez, S.; et al. Production of microbial secondary metabolites: Regulation by the carbon source. Crit. Rev. Microbiol. 2010, 36, 146–167.
- Anderson, A.J.; Dawes, E.A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 1990, 54, 450–472.
- O’Brien, J.; Wright, G.D. An ecological perspective of microbial secondary metabolism. Curr. Opin. Biotechnol. 2011, 22, 552–558.
- Deshmukh, S.K.; Prakash, V.; Ranjan, N. Marine fungi: A source of potential anticancer compounds. Front. Microbiol. 2018, 8, 2536.
- Zhu, C.; Nomura, C.T.; Perrotta, J.A.; Stipanovic, A.J.; Nakas, J.P. Production and characterization of poly-3-hydroxybutyrate from biodiesel-glycerol by Burkholderia cepacia ATCC 17759. Biotechnol. Prog. 2010, 26, 424–430.
- Lee, E.Y. Methanotrophs: Microbiology Fundamentals and Biotechnological Applications; Lee, E.Y., Ed.; Springer Nature Switzerland AG: Cham, Switzerland, 2019; ISBN 9783030232603.
- Valappil, S.P.; Misra, S.K.; Boccaccini, A.; Roy, I. Biomedical applications of polyhydroxyalkanoates, an overview of animal testing and in vivo responses. Expert Rev. Med. Devices 2006, 3, 853–868.
- Overy, D.; Rämä, T.; Oosterhuis, R.; Walker, A.; Pang, K.-L. The Neglected Marine Fungi, Sensu stricto, and Their Isolation for Natural Products’ Discovery. Mar. Drugs 2019, 17, 42.
- Puyol, D.; Batstone, D.J.; Hülsen, T.; Astals, S.; Peces, M.; Krömer, J.O. Resource recovery from wastewater by biological technologies: Opportunities, challenges, and prospects. Front. Microbiol. 2017, 7, 1–23.
- Kehrein, P.; van Loosdrecht, M.; Osseweijer, P.; Garfí, M.; Dewulf, J.; Posada, J. A critical review of resource recovery from municipal wastewater treatment plants—market supply potentials, technologies and bottlenecks. Environ. Sci. Water Res. Technol. 2020, 6, 877–910.
- Numberger, D.; Ganzert, L.; Zoccarato, L.; Mühldorfer, K.; Sauer, S.; Grossart, H.P.; Greenwood, A.D. Characterization of bacterial communities in wastewater with enhanced taxonomic resolution by full-length 16S rRNA sequencing. Sci. Rep. 2019, 9, 1–14.
- Osunmakinde, C.O.; Selvarajan, R.; Mamba, B.B.; Msagati, T.A.M. Profiling bacterial diversity and potential pathogens in wastewater treatment plants using high-throughput sequencing analysis. Microorganisms 2019, 7, 506.
- Inagaki, F.; Tsunogai, U.; Suzuki, M.; Kosaka, A.; Machiyama, H.; Takai, K.; Nunoura, T.; Nealson, K.H.; Horikoshi, K. Characterization of C 1 -Metabolizing Prokaryotic Communities in Methane Seep Habitats at the Kuroshima Knoll, Southern 16S rRNA Genes. Appl. Environ. Microbiol. 2004, 70, 7445–7455.
- Xin, J.; Zhang, Y.; Zhang, S.; Xia, C.; Li, S. Methanol production from CO2 by resting cells of the methanotrophic bacterium Methylosinus trichosporium IMV 3011. J. Basic Microbiol. 2007, 47, 426–435.
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471.
- Vaksmaa, A.; Guerrero-Cruz, S.; van Alen, T.A.; Cremers, G.; Ettwig, K.F.; Lüke, C.; Jetten, M.S.M. Enrichment of anaerobic nitrate-dependent methanotrophic ‘Candidatus Methanoperedens nitroreducens’ archaea from an Italian paddy field soil. Appl. Microbiol. Biotechnol. 2017, 101, 7075–7084.
- Jiang, H.; Chen, Y.; Jiang, P.; Zhang, C.; Smith, T.J.; Murrell, J.C.; Xing, X.-H. Methanotrophs: Multifunctional bacteria with promising applications in environmental bioengineering. Biochem. Eng. J. 2010, 49, 277–288.
- Yang, S.; Matsen, J.B.; Konopka, M.; Green-Saxena, A.; Clubb, J.; Sadilek, M.; Orphan, V.J.; Beck, D.; Kalyuzhnaya, M.G. Global Molecular Analyses of Methane Metabolism in Methanotrophic Alphaproteobacterium, Methylosinus trichosporium OB3b. Part II. Metabolomics and 13C-Labeling Study. Front. Microbiol. 2013, 4, 70.
- Whittenbury, R.; Dalton, H. The Methylotrophic Bacteria. In The Prokaryotes; Starr, M.P., Stolp, H., Trüper, H.G., Balows, A., Schlegel, H.G., Eds.; Springer: Berlin, Heidelberg, 1981; pp. 894–902. ISBN 978-3-662-13187-9.
- Kalyuzhnaya, M.G.; Khmelenina, V.N.; Kotelnikova, S.; Holmquist, L.; Pedersen, K.; Trotsenko, Y.A. Methylomonas scandinavica sp, nov., a new methanotrophic psychrotrophic bacterium isolated from deep igneous rock ground water of Sweden. Syst. Appl. Microbiol. 1999, 22, 565–572.
- Knief, C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 2015, 6, 1346.
- Strong, P.J.; Xie, S.; Clarke, W.P. Methane as a resource: Can the methanotrophs add value? Environ. Sci. Technol. 2015, 49, 4001–4018.
- AlSayed, A.; Fergala, A.; Eldyasti, A. Sustainable biogas mitigation and value-added resources recovery using methanotrophs intergrated into wastewater treatment plants. Rev. Environ. Sci. Biotechnol. 2018, 17, 351–393.
- Mahmoud, A.M.A. Biological Conversion Process of Methane into Methanol Using Mixed Culture Methanotrophic Bacteria Enriched from Activated Sludge System; York University: Toronto, ON, Canada, 2017.
- Forster, D.; Dolan, J.R.; Dunthorn, M.; Bass, D.; Bittner, L.; Boutte, C.; Christen, R.; Claverie, J.; Decelle, J.; Edvardsen, B.; et al. Benthic protists: The under-charted majority. FEMS Microbiol. Ecol. 2016, 92, 1–11.
- Kevbrina, M.V.; Okhapkina, A.A.; Akhlynin, D.S.; Kravchenko, I.K.; Nozhevnikova, A.N.; Gal’chenko, V.F.; Gal’chenko, V.F. Growth of Mesophilic Methanotrophs at Low Temperatures. Microbiology 2001, 70, 384–391.
- Liebner, S.; Wagner, D. Abundance, distribution and potential activity of methane oxidizing bacteria in permafrost soils from the Lena Delta, Siberia. Environ. Microbiol. 2007, 9, 107–117.
- Wagner, D.; Lipski, A.; Embacher, A.; Gattinger, A. Methane fluxes in permafrost habitats of the Lena Delta: Effects of microbial community structure and organic matter quality. Environ. Microbiol. 2005, 7, 1582–1592.
- Wartiainen, I.; Hestnes, A.G.; McDonald, I.R.; Svenning, M.M. Methylobacter tundripaludum sp. nov., a methane-oxidizing bacterium from Arctic wetland soil on the Svalbard islands, Norway (78° N). Int. J. Syst. Evol. Microbiol. 2006, 56, 109–113.
- Ross, M.O.; Rosenzweig, A.C. A tale of two methane monooxygenases. J. Biol. Inorg. Chem. 2017, 22, 307–319.
- Korotkova, N.; Lidstrom, M.E.; Chistoserdova, L. Identification of genes involved in the glyoxylate regeneration cycle in Methylobacterium extorquens AM1, including two new genes, meaC and meaD. J. Bacteriol. 2005, 187, 1523–1526.
- Borjesson, G.; Sundh, I.; Svensson, B. Microbial oxidation of CH4 at different temperatures in landfill cover soils. FEMS Microbiol. Ecol. 2004, 48, 305–312.
- Cui, J.; Good, N.M.; Hu, B.; Yang, J.; Wang, Q.; Sadilek, M.; Yang, S. Metabolomics Revealed an Association of Metabolite Changes and Defective Growth in Methylobacterium extorquens AM1 Overexpressing ecm during Growth on Methanol. PLoS ONE 2016, 11, e0154043.
- Korotkova, N.; Lidstrom, M.E. Connection between poly-β-hydroxybutyrate biosynthesis and growth on C1 and C2 compounds in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 2001, 183, 1038–1046.
- Babel, W. Pecularities of methylotrophs concerning overflow metabolism, especially the synthesis of polyhydroxyalkanoates. FEMS Microbiol. Lett. 1992, 103, 141–148.
This entry is offline, you can click here to edit this entry!
