Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Agriculture, Dairy & Animal Science
The virulence factors and pathogenesis of S. suis are inconclusive. Here, the associated proteins, or their derived peptides, involved in the survival of S. suis when simulated with a blood environment are demonstrated. The results reveal the derived peptides or proteins of S. suis potentially serving as the putative virulence factors. Further studies based on our findings could be used to fulfill the knowledge gap of S. suis pathogenesis.
- peptidomics analysis
- virulent peptides
- Streptococcus suis
- pathogenesis
- virulence factor
- zoonosis
1. Introduction
Streptococcus suis is a zoonotic bacterium causing severe streptococcal infection in both humans and pigs. Among 35 serotypes classified by the immunogenics of their capsular polysaccharides [1], SS serotype 2 (SS2) is the most prevalent, followed by SS14, the causative agent of S. suis zoonotic infection in humans and pigs worldwide [2][3]. For swine infections, SS7 and SS9 are associated with the clinical swine cases in the European region while SS3 is usually disseminated in North America and Asia [2]. The clinical symptoms of S. suis infection range from a common bacterial infection to streptococcal toxic shock-like syndrome (TSST) [1][4]. Although some virulence factors of S. suis such as capsular polysaccharide (CPS), suilysin, muramidase-released protein (MRP), and extracellular factors have been described [5][6][7], a thorough list of virulence factors of S. suis particularly from some other serotypes also needs to be explored. Proteomic and peptidomic techniques have been widely adopted and used to investigate the microbial proteomes for the virulence factor, diagnostic marker, and vaccine targets [8][9]. In the present study, a high-performance liquid chromatography-mass spectrometry/mass spectrometry method (LC-MS/MS) was used to determine the peptidomes of the highly virulent to humans SS2 and SS14, swine causative serotypes SS7 and SS9, and rarely reported serotypes SS25 and SS27 cultured in a specified culture medium containing whole blood, simulating their natural host environment. This study aimed to explore some putative virulence factors involved in the survival of S. suis in the blood environment.
2. Peptidomics Analysis of Virulent Peptides Involved in Streptococcus suis Pathogenesis
S. suis pathogenesis is triggered by exposure to a certain condition in the host environment [10]. In the present study, six S. suis serotypes were simulated under the human cardiovascular system and they interacted with erythrocytes, leukocytes, and other blood components particularly during bacterial septicemia (bacteremia). A large number of peptides were identified in the peptidome of S. suis. Among peptides expressed by reacting with the whole blood, only 22 unique peptides derived from S. suis proteins with known functions, matched in the STITCH database, were selected (Table 1). Interestingly, derived peptides of the ABC-type phosphate transport system and 30S ribosomal protein S2 were simultaneously co-expressed in the highly virulent SS2 and virulent SS14 cultured under the host-simulated condition. Notably, the ABC transporter protein has been reported in the pathogenicity of S. pneumoniae [11], while 4-α-glucanotransferase (malQ gene), responsible for starch metabolism in S. mutans, was expressed uniquely in the virulent SS14 [12]. Therefore, derived peptides of the ABC-type phosphate transport system, 30S ribosomal protein S2 and 4-α-glucanotransferase might serve as specific virulence factors of SS2 and SS14.
Table 1. Identified peptides derived from S. suis proteins with known function cultured under the host simulated environment.
| Gene Locus | Annotated Protein Function (Amino-Acid Length) | Function or Subcellular Location |
|---|---|---|
| SSU05_2116 | 5′-nucleotidase/2′,3′-cyclic phosphodiesterase and related esterases (448) | nucleotide catabolic activity |
| SSU05_2039 | transcriptional regulator (149) | DNA-binding transcription factor activity |
| N/A | α-1,4 glucan phosphorylase (755) | carbohydrate metabolism |
| SSU05_1489 | K01223 6-phospho-beta-glucosidase [EC-3.2.1.86] (474) |
carbohydrate metabolism |
| SSU05_0392 | 4-α-glucanotransferase (364) | carbohydrate metabolism |
| SSU05_1803 | 3-oxoacyl-[acyl-carrier-protein] reductase (244) | fatty acid biosynthesis |
| SSU05_2063 | PTS system ascorbate-specific transporter subunit IIC (482) | cell membrane/membrane |
| SSU05_1254 | ABC transporter permease (197) | transmembrane transporter |
| SSU05_1106 | ABC-type phosphate transport system (83) | periplasmic component |
| SSU05_0780 | branched-chain amino acid permease (188) | cell membrane/membrane |
| gmk | guanylate kinase (216) | GMP recycling |
| tetM | TetM (639) | response to antibiotic |
| groS | 10 kDa chaperonin (102) | ATPase activity |
| SSU05_0642 | putative low-temperature requirement A (371) | transmembrane protein |
| cps2J | Cps2J (332) | capsular biosynthesis |
| mrp | truncated MRP (1261) | secreted protein |
| rpsB | 30S ribosomal protein S2 (258) | ribosomal protein |
| SSU05_1457 | hypothetical protein (91) | |
| SSU05_0461 | hypothetical protein (567) | |
| SSU05_1869 | hypothetical protein (178) | |
| SSU05_0141 | hypothetical protein (318) | |
| SSU05_1991 | hypothetical protein (412) |
The association of 5′-nucleotidase/2′,3′-cyclic phosphodiesterase and related esterases (SSU05_0456) with S. suis has been rarely mentioned to the best of our knowledge. However, 5′-nucleotidase has been considered as a virulence factor in many pathogens including S. suis [13][14][15][16], because this enzyme is commonly involved in adenosine synthesis. Adenosine, an immunomodulatory molecule, reduces the phagocytic activity of macrophages and also impairs the neutrophil degranulation thus decreasing the inflammatory responses of the host immune system [13]. A previous study by Liu et al., (2014) demonstrated that 5′-nucleotidase is an essential factor for S. suis escaping from the host’s innate immunity as the enzyme-deficient mutant strain of S. suis lost its infectivity in the pig model [14]. Thus, our result was in concordance with Liu et al., (2014) as a derived peptide of 5′-nucleotidase was found in the virulent SS2 and SS14 reacting with the blood in the host-simulated environment. This emphasized the significance of the survival proteins and/or peptides initially expressed to tackle the host-simulated immunity.
The α-1,4 glucan phosphorylase, K01223 6-phospho-β-glucosidase [EC-3.2.1.86] and the 4-α-glucanotransferase associated with carbohydrate metabolism were exhibited in SS9 and SS25; SS2, SS14, and SS9; and SS14, respectively. These results are compatible with a study by Zhang et al., (2014) that identified 6-phospho-β-glucosidase in two Chinese isolates of virulent SS2 [17]. Unfortunately to the best of our knowledge, the direct association of these three enzymes with S. suis is not evident. However, the carbohydrate availability is positively correlated with the virulence of S. suis using the host cell invasion i.e., the adequate carbohydrate supply upregulates the virulence gene regulation of virulent S. suis as described previously in a transcriptomic study [18].
The biosynthesis of fatty acids is important for the pathogenicity and adaptation of bacteria in the host environment since fatty acids serve as an energy source and a material for cell-wall synthesis. In addition, the pattern of the lipid cell walls also acts as a factor modulating the immune responses of the host [19][20]. In the present study, a derived peptide of 3-oxoacyl-[acyl-carrier-protein] reductase responsible for the fatty acid biosynthesis was identified in SS2, SS14, SS7, and SS27. The previous studies demonstrated that this enzyme was associated with the fatty acid biosynthesis of virulent SS2 [21][22]. Thus, we could assume that this fatty acid biosynthesis-associated enzyme is significant for S. suis adaptation in the host-simulated environment.
Derived peptides of the ABC transporter permease and the ABC-type phosphate transport system were found in the peptidomes of SS14 and SS7, and SS2 and SS14, respectively. Generally, ABC transporter proteins play an important role in the transportation of various substrates including nutrients for the biosynthesis of the bacterial cell. Some of the ABC transporters serve as efflux pumps. Previous studies reported that the increased expression of an ABC efflux pump was related to fluoroquinolones resistance in S. pneumoniae and S. suis [23][24]. As shown in Figure 1, the networks of protein–protein interactions analyzed by the STITCH program, version 5.0, demonstrated high associations of the ABC-type phosphate transport system (SSU05_1106) with other phosphate ABC transporters (edge confidence score 0.900), responsible for phosphate import. Furthermore, these proteins are associated with the biofilm formation of SS2 [25] and are required for the disease pathogenesis of S. pneumoniae [11]. The branched-chain amino acid permease, one of the ABC transporter proteins, is involved in the transport of leucine, isoleucine, and valine [26]. A previous report also suggested that the branched-chain amino acid permease was related to the pathogenicity of some bacteria [11]. A certain role for the ABC transporter as a virulence factor of bacteria is still not clear. However, the presence of ABC transporters in the virulent to humans SS2, SS14, and non-virulent to humans SS7 indicated that the translocation of substrates particularly phosphate and some amino acids (leucine, isoleucine, and valine) using these ABC transporter proteins is perhaps required for the survival and then the pathogenesis (pathway) of virulent S. suis in the host-simulated environment.
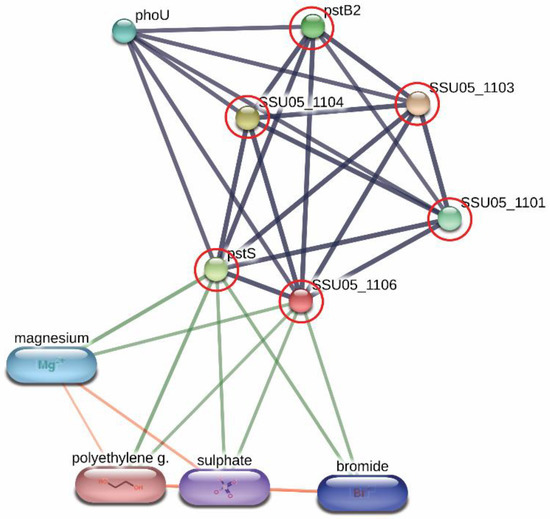
Figure 1. Relationship between ABC-type phosphate transport system (SSU05_1106) and other phosphate ABC transporters; SSU05_1101, SSU05_1103, SSU05_1104, pstS and pstB2 involved in phosphate import from STITCH 5.0 software. The grey thick lines represent the highest edge confidence scores (0.900) indicating the high strength of the protein interactions at the functional level.
Guanylate kinase phosphorylates the guanosine monophosphate to guanosine diphosphate, a precursor of the guanosine triphosphate required for the transcription of RNA synthesis. This enzyme was inhibited when the bacteria were forced to survive in the amino-acid restricted condition [27]. In the present study with adequate nutrients in the host-simulated environment, a peptide of this enzyme was identified in SS2, SS14, SS9, SS25, and SS27. This finding was in line with a previous report indicating that this enzyme is conserved and adaptively expressed depending on the nutrient availability [27].
The tetracycline-resistance gene, tetM, encoded TetM protein responsible for the inhibitory effect of tetracycline, is distributed among virulent S. suis. [28][29][30]. An outbreak report in China demonstrated that the tetracycline-resistance strains were associated with S. suis infections in both pigs and humans [28]. Our results revealed the expression of the TetM protein in SS14, SS25, and SS27 cultured under a host-simulated environment. This finding is in concordance with a previous report indicating that S. suis carried and spread the tetM gene among S. suis strains [28].
The association of S. suis and 10 kDa chaperonin is rarely reported. This protein is specified as a protein folding facilitator and also a virulence factor of bone resorption in Mycobacterium tuberculosis infection [31]. In the present study, a peptide derived from 10 kDa chaperonin was detected in the peptidome of SS7.
So far, the CPS of S. suis has been mentioned as an important virulence factor of S. suis [7]. The cps gene cluster facilitates the regulation of CPS synthesis. The Cps2J is essential for CPS biosynthesis because the deletion mutant of ∆cps2J results in the imperfection of CPS [32]. The presence of Cps2J only in SS25 might explain why the bacterium was likely to synthesize CPS to protect itself from phagocytosis by the host immune cells [7]. Cps2J was identified in SS14 and SS25 while Cps2A and Cps2C were identified in SS2, SS14, SS9, and SS25 (data not shown).
Previously, MRP has been considered as a virulent marker of S. suis [33]. The mutant strain of S. suis demonstrated its virulence even when the MRP protein was deficient. Thus, MRP is more likely to be the virulent maker than the virulence factor. This protein was proposed to be coincidentally expressed together with the true virulence factor [10]. In this study, we identified a peptide derived from truncated MRP only in SS7 under the host-simulated environment. The limited existence of this protein might be explained by the fact that MRP is a cell-wall anchored protein and is released into the supernatant part of the culture medium. Our peptide preparation method did not precipitate proteins in the supernatant thus decreasing the chance of capturing MRP in S. suis cells. A previous study focusing on proteomic analysis of SS2 human isolates also reported no expression of MRP either [34].
Other unique proteins identified in this study were putative low-temperature requirement A protein (SSU05_0642), transcriptional regulator (SSU05_2039), and 30S ribosomal protein S2(rpsB). Although the related information regarding their virulence is not available, the expression of 30S ribosomal protein S2 should not be neglected, since these proteins were exhibited only in virulent SS2 and SS14 culture under the host-simulated environment. The role of these proteins in conjunction with the pathogenesis of virulent S. suis deserves further study.
Among five hypothetical proteins presented in this study (Table 1), SSU05_1869 hypothetical protein was previously mentioned as a putative ABC transporter protein [35] and SSU05_1991 hypothetical protein was described as possibly associated with bacterial cell growth and division in S. suis serotype 2 [36]. In the present study, SSU05_1991 hypothetical protein was also exhibited in SS2 and also in SS7, SS25, and SS27. When the functions or subcellular locations of the other three hypothetical proteins SSU05_1457, SSU05_0461, and SSU05_0141 are identified, their roles regarding the pathogenesis of virulent S. suis will be disclosed.
3. Conclusions
S. suis expressed its virulence factor in the form of proteins or peptides to survive in the host. Our study demonstrated various derived peptides expressed in S. suis SS2, SS14, SS7, SS9, SS25, and SS27 reacting with whole blood. Most unique proteins were associated with the transcriptional process and cellular metabolic processes, thus indicating the adaptation of the bacterial cell during the initial period of incubation time in the host-simulated environment. The peptides of the ABC-type phosphate transport system (SSU05_1106) and 30S ribosomal protein S2 (rpsB) were solely expressed in virulent to humans SS2 and SS14 under the host-simulated environment and are proposed as putative virulence factors potentially involved in S. suis pathogenesis. The results of this study apply to the diagnostic marker and vaccine targets [8][9]. However, our results only demonstrated the identified peptides by in vitro simulating environment. Further study by in vivo and gene knockout should be conducted to verify our peptidomic findings.
This entry is adapted from the peer-reviewed paper 10.3390/ani11092480
References
- Staats, J.J.; Feder, I.; Okwumabua, O.; Chengappa, M.M. Streptococcus suis: Past and present. Vet. Res. Commun. 1997, 21, 381–407.
- Goyette-Desjardins, G.; Auger, J.P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014, 3, e45.
- Gajdacs, M.; Nemeth, A.; Knausz, M.; Barrak, I.; Stajer, A.; Mestyan, G.; Melegh, S.; Nyul, A.; Toth, A.; Agoston, Z.; et al. Streptococcus suis: An Underestimated Emerging Pathogen in Hungary? Microorganisms 2020, 8, 1292.
- Tang, J.; Wang, C.; Feng, Y.; Yang, W.; Song, H.; Chen, Z.; Yu, H.; Pan, X.; Zhou, X.; Wang, H.; et al. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 2006, 3, e151.
- Baums, C.G.; Valentin-Weigand, P. Surface-associated and secreted factors of Streptococcus suis in epidemiology, pathogenesis and vaccine development. Anim. Health Res. Rev. 2009, 10, 65–83.
- Jacobs, A.A.; Loeffen, P.L.; van den Berg, A.J.; Storm, P.K. Identification, purification, and characterization of a thiol-activated hemolysin (suilysin) of Streptococcus suis. Infect. Immun. 1994, 62, 1742–1748.
- Smith, H.E.; Damman, M.; van der Velde, J.; Wagenaar, F.; Wisselink, H.J.; Stockhofe-Zurwieden, N.; Smits, M.A. Identification and characterization of the cps locus of Streptococcus suis serotype 2: The capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 1999, 67, 1750–1756.
- Perez-Llarena, F.J.; Bou, G. Proteomics as a Tool for Studying Bacterial Virulence and Antimicrobial Resistance. Front. Microbiol. 2016, 7, 410.
- Wu, H.J.; Wang, A.H.J.; Jennings, M.P. Discovery of virulence factors of pathogenic bacteria. Curr. Opin. Chem. Biol. 2008, 12, 93–101.
- Fittipaldi, N.; Segura, M.; Grenier, D.; Gottschalk, M. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 2012, 7, 259–279.
- Basavanna, S.; Khandavilli, S.; Yuste, J.; Cohen, J.M.; Hosie, A.H.; Webb, A.J.; Thomas, G.H.; Brown, J.S. Screening of Streptococcus pneumoniae ABC transporter mutants demonstrates that LivJHMGF, a branched-chain amino acid ABC transporter, is necessary for disease pathogenesis. Infect. Immun. 2009, 77, 3412–3423.
- Sato, Y.; Okamoto-Shibayama, K.; Azuma, T. The malQ gene is essential for starch metabolism in Streptococcus mutans. J. Oral Microbiol. 2013, 5, 21285.
- Zheng, L.; Khemlani, A.; Lorenz, N.; Loh, J.M.; Langley, R.J.; Proft, T. Streptococcal 5′-Nucleotidase A (S5nA), a Novel Streptococcus pyogenes Virulence Factor That Facilitates Immune Evasion. J. Biol. Chem. 2015, 290, 31126–31137.
- Liu, P.; Pian, Y.; Li, X.; Liu, R.; Xie, W.; Zhang, C.; Zheng, Y.; Jiang, Y.; Yuan, Y. Streptococcus suis adenosine synthase functions as an effector in evasion of PMN-mediated innate immunit. J. Infect. Dis. 2014, 210, 35–45.
- Thammavongsa, V.; Kern, J.W.; Missiakas, D.M.; Schneewind, O. Staphylococcus aureus synthesizes adenosine to escape host immune responses. J. Exp. Med. 2009, 206, 2417–2427.
- Fan, J.; Zhang, Y.; Chuang-Smith, O.N.; Frank, K.L.; Guenther, B.D.; Kern, M.; Schlievert, P.M.; Herzberg, M.C. Ecto-5′-nucleotidase: A candidate virulence factor in Streptococcus sanguinis experimental endocarditis. PLoS ONE 2012, 7, e38059.
- Zhang, D.; Du, N.; Ma, S.; Hu, Q.; Lu, G.; Chen, W.; Zeng, C. In vitro transcriptome analysis of two Chinese isolates of Streptococcus suis serotype 2. Genom. Proteom. Bioinform. 2014, 12, 266–275.
- Ferrando, M.L.; van Baarlen, P.; Orru, G.; Piga, R.; Bongers, R.S.; Wels, M.; De Greeff, A.; Smith, H.E.; Wells, J.M. Carbohydrate availability regulates virulence gene expression in Streptococcus suis. PLoS ONE 2014, 9, e89334.
- Rameshwaram, N.R.; Singh, P.; Ghosh, S.; Mukhopadhyay, S. Lipid metabolism and intracellular bacterial virulence: Key to next-generation therapeutics. Future Microbiol. 2018, 13, 1301–1328.
- Ghazaei, C. Mycobacterium tuberculosis and lipids: Insights into molecular mechanisms from persistence to virulence. J. Res. Med. Sci. 2018, 23, 63.
- Gao, T.; Yuan, F.; Liu, Z.; Liu, W.; Zhou, D.; Yang, K.; Duan, Z.; Guo, R.; Liang, W.; Hu, Q.; et al. MnmE, a Central tRNA-Modifying GTPase, Is Essential for the Growth, Pathogenicity, and Arginine Metabolism of Streptococcus suis Serotype 2. Front. Cell Infect. Microbiol. 2019, 9, 173.
- Gao, T.; Yuan, F.; Liu, Z.; Liu, W.; Zhou, D.; Yang, K.; Guo, R.; Liang, W.; Zou, G.; Zhou, R.; et al. Proteomic and Metabolomic Analyses Provide Insights into the Mechanism on Arginine Metabolism Regulated by tRNA Modification Enzymes GidA and MnmE of Streptococcus suis. Front. Cell Infect. Microbiol. 2020, 10, 597408.
- Yao, J.; Shang, K.; Huang, J.; Ran, W.; Kashif, J.; Wang, L. Overexpression of an ABC transporter and mutations of GyrA, GyrB, and ParC in contributing to high-level ciprofloxacin resistance in Streptococcus suis type 2. Biosci. Trends 2014, 8, 84–92.
- El Garch, F.; Lismond, A.; Piddock, L.J.; Courvalin, P.; Tulkens, P.M.; Van Bambeke, F. Fluoroquinolones induce the expression of patA and patB, which encode ABC efflux pumps in Streptococcus pneumoniae. J. Antimicrob Chemother 2010, 65, 2076–2082.
- Yang, Y.B.; Chen, J.Q.; Zhao, Y.L.; Bai, J.W.; Ding, W.Y.; Zhou, Y.H.; Chen, X.Y.; Liu, D.; Li, Y.H. Sub-MICs of Azithromycin Decrease Biofilm Formation of Streptococcus suis and Increase Capsular Polysaccharide Content of S. suis. Front. Microbiol. 2016, 7, 1659.
- Grauslund, M.; Didion, T.; Kielland-Brandt, M.C.; Andersen, H.A. BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1995, 1269, 275–280.
- Liu, K.; Myers, A.R.; Pisithkul, T.; Claas, K.R.; Satyshur, K.A.; Amador-Noguez, D.; Keck, J.L.; Wang, J.D. Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol. Cell 2015, 57, 735–749.
- Ye, C.; Bai, X.; Zhang, J.; Jing, H.; Zheng, H.; Du, H.; Cui, Z.; Zhang, S.; Jin, D.; Xu, Y.; et al. Spread of Streptococcus suis sequence type 7, China. Emerg. Infect. Dis. 2008, 14, 787–791.
- Gurung, M.; Tamang, M.D.; Moon, D.C.; Kim, S.R.; Jeong, J.H.; Jang, G.C.; Jung, S.C.; Park, Y.H.; Lim, S.K. Molecular Basis of Resistance to Selected Antimicrobial Agents in the Emerging Zoonotic Pathogen Streptococcus suis. J. Clin. Microbiol. 2015, 53, 2332–2336.
- Nicholson, T.L.; Waack, U.; Anderson, T.K.; Bayles, D.O.; Zaia, S.R.; Goertz, I.; Eppinger, M.; Hau, S.J.; Brockmeier, S.L.; Shore, S.M. Comparative Virulence and Genomic Analysis of Streptococcus suis Isolates. Front. Microbiol. 2020, 11, 620843.
- Roberts, M.M.; Coker, A.R.; Fossati, G.; Mascagni, P.; Coates, A.R.; Wood, S.P. Mycobacterium tuberculosis chaperonin 10 heptamers self-associate through their biologically active loops. J. Bacteriol. 2003, 185, 4172–4185.
- Zhang, Y.; Ding, D.; Liu, M.; Yang, X.; Zong, B.; Wang, X.; Chen, H.; Bei, W.; Tan, C. Effect of the glycosyltransferases on the capsular polysaccharide synthesis of Streptococcus suis serotype 2. Microbiol. Res. 2016, 185, 45–54.
- Vecht, U.; Wisselink, H.J.; Jellema, M.L.; Smith, H.E. Identification of two proteins associated with virulence of Streptococcus suis type 2. Infect. Immun. 1991, 59, 3156–3162.
- Prados de la Torre, E.; Rodriguez-Franco, A.; Rodriguez-Ortega, M.J. Proteomic and Bioinformatic Analysis of Streptococcus suis Human Isolates: Combined Prediction of Potential Vaccine Candidates. Vaccines 2020, 8, 188.
- Pian, Y.; Wang, P.; Liu, P.; Zheng, Y.; Zhu, L.; Wang, H.; Xu, B.; Yuan, Y.; Jiang, Y. Proteomics identification of novel fibrinogen-binding proteins of Streptococcus suis contributing to antiphagocytosis. Front. Cell Infect. Microbiol. 2015, 5, 19.
- Gao, T.; Tan, M.; Liu, W.; Zhang, C.; Zhang, T.; Zheng, L.; Zhu, J.; Li, L.; Zhou, R. GidA, a tRNA Modification Enzyme, Contributes to the Growth, and Virulence of Streptococcus suis Serotype 2. Front. Cell Infect. Microbiol. 2016, 6, 44.
This entry is offline, you can click here to edit this entry!
