Given the global scale of the COVID-19 pandemic and the health emergency it has caused, it is crucial to understand the impact of SARS-CoV-2 and its mutations. Here, we comprehensively review SARS-CoV-2 interactions with host cells, describe SARS-CoV-2 variants, assess impact of their protein mutations and enumerate databases with SARS-CoV-2 host-pathogen interaction data.
- SARS-CoV-2
- SARS-CoV-2 Mutants
- Host Pathogen Interactions
- Proteins
- miRNA
- Innate Immunity
Introduction:
The outbreak of Coronavirus Disease 2019 (COVID-19) is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). The virus was first detected in the city of Wuhan, China, around December 2019 [1]. Due to the exponential rise in COVID-19 infections across countries, COVID-19 was declared a pandemic by the World Health Organization (WHO) on 11 March 2020 [2]. COVID-19 has caused a global health crisis, infecting over 191 million individuals with over 4 million deaths as of June 2021 (https: //www.worldometers.info/coronavirus/, accessed 30 June 2021).
SARS-CoV-2 is among the largest RNA viruses, ranging from 26-32 kilobases, and comprises two large open reading frames (ORF1a and ORF1b) [3] [4]. The two ORFs translate into replicase polypeptides (pp1a and pp1b) that form the non-structural proteins essential for viral replication [5] [6]. The complete assembly of SARS-CoV-2 is aided by the structural proteins (spike (S), envelope (E), membrane (M), and nucleoprotein (N)). [6].
RNA viruses demonstrate rapid evolution due to a high mutation rate which is a million times higher than the host mutation rate [7] [8]. The high mutation rate in SARS-CoV-2 is attributed to the enormous genome variability that enabled it to the escape host immune response and antiviral therapeutics. The evolving nature of SARS-CoV-2 has resulted in several new strains of the virus across the world, including highly infectious B.1.1.7/Alpha (UK), B.1.351/Beta (South Africa), B. 1.1.28/Gamma (Brazil), B.1.617.2/Delta (India), and C.37/Lambda (South America) variants [9] [10] [11] [12] [13]. Multiple studies have reported that rapidly evolving new strains of SARS-CoV-2 exhibit decreased susceptibility to antiviral therapeutics and escape neutralization by vaccine-induced humoral immunity in the host [14] [15] [16]. The evolution has urged for the need to dissect the molecular features in the virus that enhance its infectious capacity and modulates host cells through direct and indirect interactions in various cellular components. Several studies on sequence variation in the SARS-CoV-2 genome have also identified an abundance of mutations in the spike protein of the virus that enables its entry into the host cell [17] [18] [19] [20] [21] [22].
Results:
1 SARS-CoV-2 Interactions with Host Cellular Components
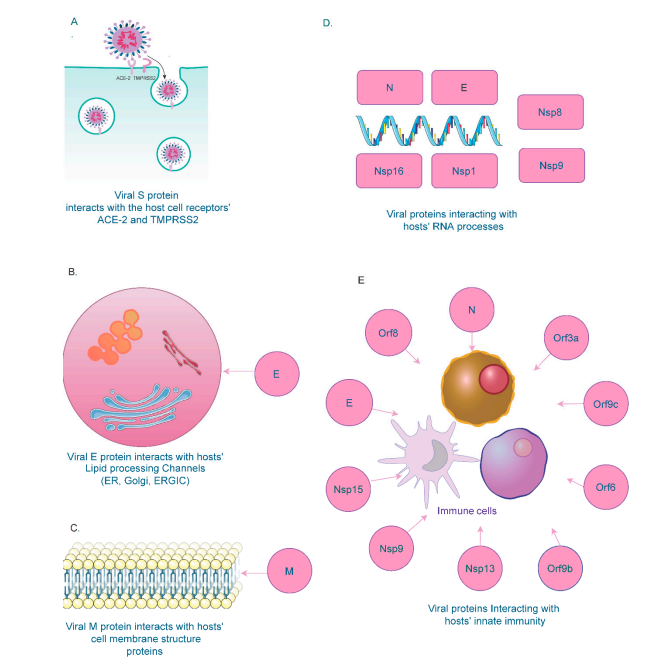
Various structural, non-structural, and accessory proteins interact with host cellular components to regulate the biological processes [23]. Several host RNA molecules such as small nuclear RNA (snRNA), 18s rRNA, and the 7SL RNA component of the signal recognition particle (SRP) are shown to interact with SARS-CoV-2 [24]. The E protein binds to BRD proteins and disrupts BRD binding with histones [25] [26]. E and M proteins are also found in cellular compartments involved in intracellular tracking (Golgi apparatus, ER, ERGIC) [27] [28]. The N protein interacts with host factors RNA factors to protect genome from nucleases and pattern recognition proteins [29]. Non-structural proteins interact with snRNAs and signal recognition proteins [30].
Figure 1: SARS-CoV-2 Interactions with Host Cellular Components
2 SARS-CoV-2 Interactions with Host Immune System
Various viral structures have been identified to interact with several components of the host innate immune system. Several of the viral non-structural proteins (Orf6, Orf9, Nsp13, and Nsp15) targets proteins of the IFN pathway, resulting in a dysregulated immune response [31] [32] [33]. Viral E and ORF8 proteins also promote high cytokine levels; cytokine storms marked by hyperactive inflammatory response are associated with severe COVID-19 cases and poor outcomes [34] [35]. Depletion of cellular nucleic acid binding proteins is also associated with higher viral titers [36].
3 SARS-CoV-2 Interactions with Host RNA-Binding Proteins & microRNAs
SARS-CoV-2 modulates the host immune response by suppressing these signaling pathways to support the viral life cycle and propagate infection. Experimental and computational studies indicate multiple host RBPs have direct and specific binding to the SARS-CoV-2 RNA genome. miRNA profiling experiments and computations also find miRNA with binding sites in SARS-CoV-2 ORFs and 5′ and 3′ UTRs also modulate mRNAs of proteins associated with viral entry [37]. Sequestration of miRNA on the viral genome could also enhance viral replication and suppress the host immune response[38] [39].
| Interactions between SARS-CoV-2, Human RBPs, and miRNAs | Technique Used | References |
|---|---|---|
| 332 SARS-CoV2–host protein–protein interaction | AP-MS | [90] |
| 309 host proteins interaction with SARS-CoV-2 RNA | ChIRP-MS | [129] |
| Host RAB2A, RAB7A, and RAB10 interaction with both viral RNA and protein | CRISPR Cas-9-based perturbation | [130] |
| 25 human RBPs targeting SARS-CoV-2 viral RNA | RBP motif-based in silico prediction | [133] |
| 104 human proteins directly and specifically bind to SARS-CoV-2 RNAs | RAP-MS | [87] |
| 288 host miRNAs predicted to bind SARS-CoV-2 (ORF1ab, N, S, 5′-UTR, and 3′-UTR) | Bioinformatic prediction algorithms and miRNA profiling | [142] |
| 479 human miRNAs could target various SARS-CoV-2 genes (S, E, M, N, Orf 1ab, 3a, 6, 7, 8, and 10) | Machine learning-based miRNA prediction | [140] |
| 22 miRNAs could bind throughout the length of the SARS-CoV-2 viral genome | Computational approach using FIMO | [133] |
4 SARS-CoV-2 Protein Mutations & Impact
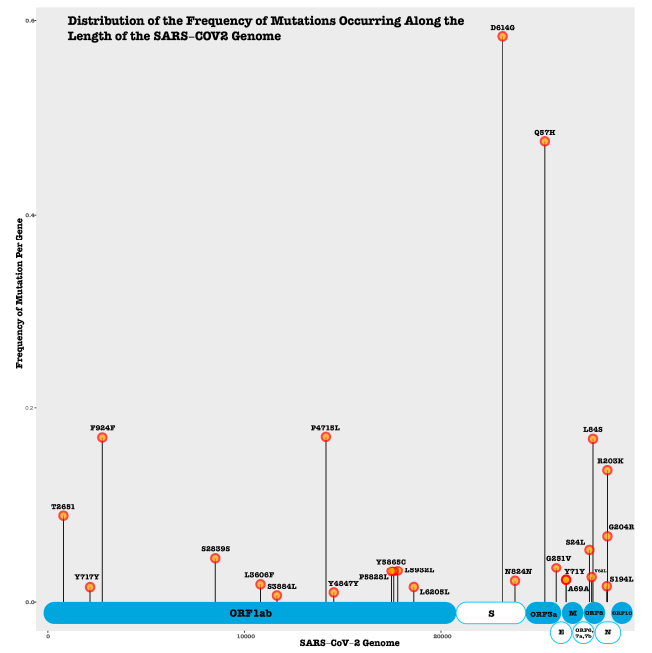
We evaluate reported and predicted synonymous, nonsynonymous, insertion, and deletion mutations in SARS-CoV-2 proteins. Many mutations occur in the spike (S) and nucleocapsid proteins (N) and ORF1ab polyprotein, which are vital for viral cellular entry, structure and replication. Several non-synonymous mutations (S N354D, N R203K) alter protein secondary structure and solvent accessibility [40] [41] . Moreover, some mutations may be linked to changes in virus transmissibility and antigenicity.
Figure 2: SARS-CoV-2 Protein Mutations
5 Database Resources for SARS-CoV-2 Host-Pathogen Interactions
Several databases provide SARS-CoV-2 Host-Pathogen Interaction (HPI) data including protein–protein, protein–RNA and RNA–RNA interactions. We identify eight public-available databases covering at least one of these interaction types: SARS-3D, VirHostnet, BioGRID, IntAct, Human Proteome Atlas, Intomics, Protein Data Bank and STRING-DB. We also note six other databases well poised to accomodate SARS-CoV-2 HPI data.
| Database | Interaction(s) | URL | Description |
|---|---|---|---|
| SARS-3D | Protein–protein | http://sars3d.com (accessed on 30 June 2021) |
3D protein models predicted using genome data |
| VirHostNet | Protein–protein | http://virhostnet.prabi.fr (accessed on 30 June 2021) |
Interactions between SARS-CoV-2 and human proteins |
| BioGRID (curated dataset) | Protein–protein | https://thebiogrid.org/project/3 (accessed on 30 June 2021) |
Curated coronavirus dataset with 22,223 interactions over 110 proteins |
| IntAct | Protein–protein and protein–RNA | https://www.ebi.ac.uk/intact/query/annot:%22dataset:coronavirus%22 (accessed on 30 June 2021) |
Over 4400 binarized SARS-CoV-2–human molecular interactions |
| Human Proteome Atlas | Protein–protein | https://www.proteinatlas.org (accessed on 30 June 2021) |
Summary of tissue and cell expression patterns of human proteins interacting with SARS-CoV-2 |
| Intomics | Protein–protein | https://www.intomics.com/covid19/?utm_source=intomics&utm_medium=linkedin&utm_campaign=covid19 (accessed on 30 June 2021) |
PPI network based on transcriptional response in human SARS-CoV-2-infected cells |
| Protein Data Bank | Protein–protein | https://www.rcsb.org/news?year=2020&article=5e74d55d2d410731e9944f52&feature=true (accessed on 30 June 2021) |
Protein–protein complex crystal structures (i.e., S—ACE2 complex) |
| STRING-DB | Protein–protein | https://string-db.org/cgi/covid.pl (accessed on 30 June 2021) |
Protein–protein interaction network with 332 virus-interacting human proteins |
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms9091794
References
- Harapan Harapan; Naoya Itoh; Amanda Yufika; Wira Winardi; Synat Keam; Haypheng Te; Dewi Megawati; Zinatul Hayati; Abram L. Wagner; Mudatsir Mudatsir; et al. Coronavirus disease 2019 (COVID-19): A literature review. Journal of Infection and Public Health 2020, 13, 667-673, 10.1016/j.jiph.2020.03.019.
- Xiaoyi Huang; Fengxiang Wei; Liang Hu; Lijuan Wen; Ken Chen; Epidemiology and Clinical Characteristics of COVID-19. Archives of Iranian Medicine 2020, 23, 268-271, 10.34172/aim.2020.09.
- Yu Chen; Qianyun Liu; Deyin Guo; Emerging coronaviruses: Genome structure, replication, and pathogenesis. Journal of Medical Virology 2020, 92, 418-423, 10.1002/jmv.25681.
- Shuo Su; Gary Wong; Weifeng Shi; Jun Liu; Alexander C.K. Lai; Jiyong Zhou; Wenjun Liu; Yuhai Bi; George F. Gao; Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends in Microbiology 2016, 24, 490-502, 10.1016/j.tim.2016.03.003.
- Azadeh Rahimi; Azin Mirzazadeh; Soheil Tavakolpour; Genetics and genomics of SARS-CoV-2: A review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics 2020, 113, 1221-1232, 10.1016/j.ygeno.2020.09.059.
- Ahmad Abu Turab Naqvi; Kisa Fatima; Taj Mohammad; Urooj Fatima; Indrakant K Singh; Archana Singh; Shaikh Muhammad Atif; Gururao Hariprasad; Gulam Mustafa Hasan; Imtaiyaz Hassan; et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 2020, 1866, 165878-165878, 10.1016/j.bbadis.2020.165878.
- Siobain Duffy; Why are RNA virus mutation rates so damn high?. PLOS Biology 2018, 16, e3000003, 10.1371/journal.pbio.3000003.
- John W. Drake; John J. Holland; Mutation rates among RNA viruses. Proceedings of the National Academy of Sciences 1999, 96, 13910-13913, 10.1073/pnas.96.24.13910.
- Ingrid Torjesen; Covid-19: Delta variant is now UK’s most dominant strain and spreading through schools. BMJ 2021, 373, n1445, 10.1136/bmj.n1445.
- Venkata-Viswanadh Edara; Lilin Lai; Malaya K. Sahoo; Katharine Floyd; Mamdouh Sibai; Daniel Solis; Maria W. Flowers; Laila Hussaini; Caroline Rose Ciric; Sarah Bechnack; et al. Infection and vaccine-induced neutralizing antibody responses to the SARS-CoV-2 B.1.617.1 variant. The New England Journal of Medicine 2021, 385, 664-666, 10.1101/2021.05.09.443299.
- Bernard La Scola; Philippe Lavrard; Pierre-Edouard Fournier; Philippe Colson; Alexandre Lacoste; Didier Raoult; SARS-CoV-2 variant from India to Marseille: The still active role of ports in the introduction of epidemics. Travel Medicine and Infectious Disease 2021, 42, 102085-102085, 10.1016/j.tmaid.2021.102085.
- Jianying Liu; Yang Liu; Hongjie Xia; Jing Zou; Scott C. Weaver; Kena A. Swanson; Hui Cai; Mark Cutler; David Cooper; Alexander Muik; et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature 2021, 596, 273-275, 10.1038/s41586-021-03693-y.
- Arnaud Fontanet; Brigitte Autran; Bruno Lina; Marie Paule Kieny; Salim S Abdool Karim; Devi Sridhar; SARS-CoV-2 variants and ending the COVID-19 pandemic. The Lancet 2021, 397, 952-954, 10.1016/s0140-6736(21)00370-6.
- Pengfei Wang; Manoj S. Nair; Lihong Liu; Sho Iketani; Yang Luo; Yicheng Guo; Maple Wang; Jian Yu; Baoshan Zhang; Peter D. Kwong; et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135, 10.1101/2021.01.25.428137.
- Wilfredo F. Garcia-Beltran; Evan C. Lam; Kerri St. Denis; Adam D. Nitido; Zeidy H. Garcia; Blake M. Hauser; Jared Feldman; Maia N. Pavlovic; David J. Gregory; Mark C. Poznansky; et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 184, 2372-2383.e9, 10.1016/j.cell.2021.03.013.
- Carmen Gómez; Beatriz Perdiguero; Mariano Esteban; Emerging SARS-CoV-2 Variants and Impact in Global Vaccination Programs against SARS-CoV-2/COVID-19. Vaccines 2021, 9, 243, 10.3390/vaccines9030243.
- Ivair José Morais; Richard Costa Polveiro; Gabriel Medeiros Souza; Daniel Inserra Bortolin; Flávio Tetsuo Sassaki; Alison Talis Martins Lima; The global population of SARS-CoV-2 is composed of six major subtypes. Scientific Reports 2020, 10, 1-9, 10.1038/s41598-020-74050-8.
- Lalitha Guruprasad; Human SARS CoV ‐2 spike protein mutations. Proteins: Structure, Function, and Genetics 2021, 89, 569-576, 10.1002/prot.26042.
- Sonia Jangra; Chengjin Ye; Raveen Rathnasinghe; Daniel Stadlbauer; Florian Krammer; Viviana Simon; Luis Martinez-Sobrido; Adolfo García-Sastre; Michael Schotsaert; PVI study group; et al. The E484K mutation in the SARS-CoV-2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post-vaccination sera. medRxiv 2021, na, na, 10.1101/2021.01.26.21250543.
- Lizhou Zhang; Cody B. Jackson; Huihui Mou; Amrita Ojha; Haiyong Peng; Brian D. Quinlan; Erumbi S. Rangarajan; Andi Pan; Abigail Vanderheiden; Mehul S. Suthar; et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nature Communications 2020, 11, 1-9, 10.1038/s41467-020-19808-4.
- Bette Korber; Will M. Fischer; Sandrasegaram Gnanakaran; Hyejin Yoon; James Theiler; Werner Abfalterer; Nick Hengartner; Elena E. Giorgi; Tanmoy Bhattacharya; Brian Foley; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812-827.e19, 10.1016/j.cell.2020.06.043.
- Sinae Kim; Jong Ho Lee; Siyoung Lee; Saerok Shim; Tam T. Nguyen; JiHyeong Hwang; Heijun Kim; Yeo-Ok Choi; Jaewoo Hong; Suyoung Bae; et al. The Progression of SARS Coronavirus 2 (SARS-CoV2): Mutation in the Receptor Binding Domain of Spike Gene. Immune Network 2019, 20, e41, 10.4110/in.2020.20.e41.
- Emanuel Wyler; Kirstin Mösbauer; Vedran Franke; Asija Diag; Lina Theresa Gottula; Roberto Arsiè; Filippos Klironomos; David Koppstein; Katja Hönzke; Salah Ayoub; et al. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience 2021, 24, 102151-102151, 10.1016/j.isci.2021.102151.
- Denis L.J. Lafontaine; David Tollervey; The function and synthesis of ribosomes. Nature Reviews Molecular Cell Biology 2001, 2, 514-520, 10.1038/35080045.
- Shitao Li; Lingyan Wang; Michael Berman; Young-Yun Kong; Martin E. Dorf; Mapping a Dynamic Innate Immunity Protein Interaction Network Regulating Type I Interferon Production. Immunity 2011, 35, 426-440, 10.1016/j.immuni.2011.06.014.
- J C Hagege; [Rhinoplasty before and after--18 and 40 years].. Annales de Chirurgie Plastique Esthétique 1987, 33, na, .
- He-Wei Jiang; Hai-Nan Zhang; Qing-Feng Meng; Jia Xie; Yang Li; Hong Chen; Yun-Xiao Zheng; Xue-Ning Wang; Huan Qi; Jing Zhang; et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cellular & Molecular Immunology 2020, 17, 998-1000, 10.1038/s41423-020-0514-8.
- Jingjiao Li; Mingquan Guo; Xiaoxu Tian; Xin Wang; Xing Yang; Ping Wu; Chengrong Liu; Zixuan Xiao; Yafei Qu; Yue Yin; et al. Virus-Host Interactome and Proteomic Survey Reveal Potential Virulence Factors Influencing SARS-CoV-2 Pathogenesis. Med 2020, 2, 99-112.e7, 10.1016/j.medj.2020.07.002.
- Theodora Myrto Perdikari; Anastasia C. Murthy; Veronica H. Ryan; Scott Watters; Mandar T. Naik; Nicolas L. Fawzi; SARS‐CoV‐2 nucleocapsid protein phase‐separates with RNA and with human hnRNPs. The EMBO Journal 2020, 39, e106478, 10.15252/embj.2020106478.
- Abhik K. Banerjee; Mario R. Blanco; Emily A. Bruce; Drew D. Honson; Linlin M. Chen; Amy Chow; Prashant Bhat; Noah Ollikainen; Sofia A. Quinodoz; Colin Loney; et al. SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses. Cell 2020, 183, 1325-1339.e21, 10.1016/j.cell.2020.10.004.
- Joachim L. Schultze; Anna C. Aschenbrenner; COVID-19 and the human innate immune system. Cell 2021, 184, 1671-1692, 10.1016/j.cell.2021.02.029.
- Lisa Miorin; Thomas Kehrer; Maria Teresa Sanchez-Aparicio; Ke Zhang; Phillip Cohen; Roosheel S. Patel; Anastasija Cupic; Tadashi Makio; Menghan Mei; Elena Moreno; et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proceedings of the National Academy of Sciences 2020, 117, 28344-28354, 10.1073/pnas.2016650117.
- Hongjie Xia; Zengguo Cao; Xuping Xie; Xianwen Zhang; John Yun-Chung Chen; Hualei Wang; Vineet D. Menachery; Ricardo Rajsbaum; Pei-Yong Shi; Evasion of Type I Interferon by SARS-CoV-2. Cell Reports 2020, 33, 108234-108234, 10.1016/j.celrep.2020.108234.
- Dina Ragab; Haitham Salah Eldin; Mohamed Taeimah; Rasha Khattab; Ramy Salem; The COVID-19 Cytokine Storm; What We Know So Far. Frontiers in Immunology 2020, 11, 1446, 10.3389/fimmu.2020.01446.
- Guang Chen; Di Wu; Wei Guo; Yong Cao; Da Huang; Hongwu Wang; Tao Wang; Xiaoyun Zhang; Huilong Chen; Haijing Yu; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. Journal of Clinical Investigation 2020, 130, 2620-2629, 10.1172/jci137244.
- Nora Schmidt; Caleb A. Lareau; Hasmik Keshishian; Sabina Ganskih; Cornelius Schneider; Thomas Hennig; Randy Melanson; Simone Werner; Yuanjie Wei; Matthias Zimmer; et al. The SARS-CoV-2 RNA–protein interactome in infected human cells. Nature Microbiology 2020, 6, 339-353, 10.1038/s41564-020-00846-z.
- Jacob B. Pierce; Viorel Simion; Basak Icli; Daniel Pérez-Cremades; Henry S. Cheng; Mark W. Feinberg; Computational Analysis of Targeting SARS-CoV-2, Viral Entry Proteins ACE2 and TMPRSS2, and Interferon Genes by Host MicroRNAs. Genes 2020, 11, 1354, 10.3390/genes11111354.
- Rajneesh Srivastava; Swapna Daulatabad; Mansi Srivastava; Sarath Janga; Role of SARS-CoV-2 in Altering the RNA-Binding Protein and miRNA-Directed Post-Transcriptional Regulatory Networks in Humans. International Journal of Molecular Sciences 2020, 21, 7090, 10.3390/ijms21197090.
- Rafal Bartoszewski; Michal Dabrowski; Bogdan Jakiela; Sadis Matalon; Kevin S. Harrod; Marek Sanak; James F. Collawn; SARS-CoV-2 may regulate cellular responses through depletion of specific host miRNAs. American Journal of Physiology-Lung Cellular and Molecular Physiology 2020, 319, L444-L455, 10.1152/ajplung.00252.2020.
- Thanh Thi Nguyen; Pubudu N. Pathirana; Thin Nguyen; Quoc Viet Hung Nguyen; Asim Bhatti; Dinh C. Nguyen; Dung Tien Nguyen; Ngoc Duy Nguyen; Douglas Creighton; Mohamed Abdelrazek; et al. Genomic mutations and changes in protein secondary structure and solvent accessibility of SARS-CoV-2 (COVID-19 virus). Scientific Reports 2021, 11, 1-16, 10.1038/s41598-021-83105-3.
- Siqi Wu; Chang Tian; Panpan Liu; Dongjie Guo; Wei Zheng; Xiaoqiang Huang; Yang Zhang; Lijun Liu; Effects of SARS‐CoV‐2 mutations on protein structures and intraviral protein–protein interactions. Journal of Medical Virology 2020, 93, 2132-2140, 10.1002/jmv.26597.
