Poly (ADP-ribose) polymerases (PARP) 1-3 are well-known multi-domain enzymes, catalysing the covalent modification of proteins, DNA, and themselves. They attach mono- or poly-ADP-ribose to targets using NAD+ as a substrate. Poly-ADP-ribosylation (PARylation) is central to the important functions of PARP enzymes in the DNA damage response and nucleosome remodelling. Activation of PARP happens through DNA binding via zinc fingers and/or the WGR domain. Modulation of their activity using PARP inhibitors occupying the NAD+ binding site has proven successful in cancer therapies. For decades, studies set out to elucidate their full-length molecular structure and activation mechanism.
- Poly (ADP-ribose) polymerases 1-3
- DNA damage response
- PARP-DNA binding
- PARP activation
1. Introduction
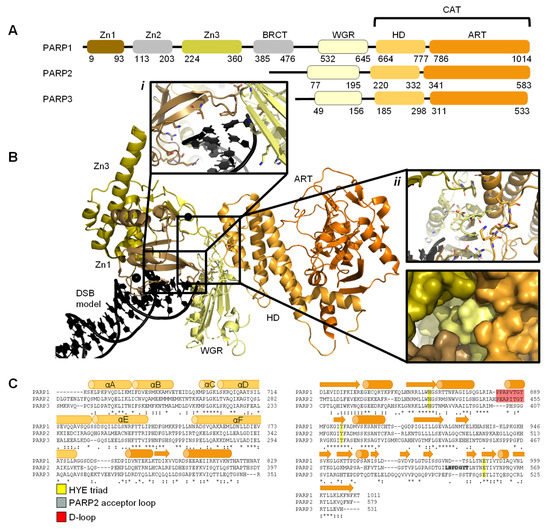
2. DNA Damage Recognition by PARP Enzymes
2.1. PARP1 Zn Fingers Bind a DNA Break
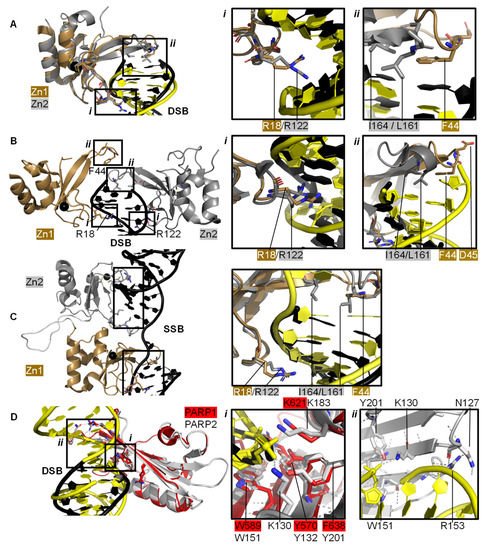
2.2. Recognition and Binding of Other DNA Breaks via PARP Domains
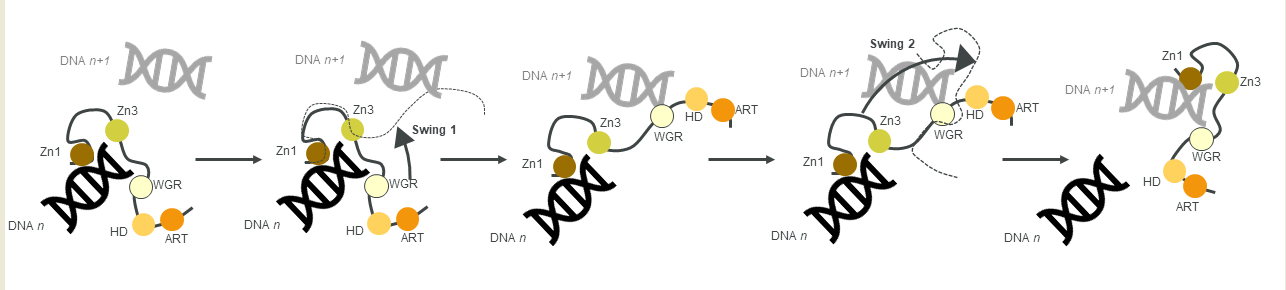
2.3. The PARP Paradox: Finding a Needle in a Haystack
This entry is adapted from the peer-reviewed paper 10.3390/ijms22105112
References
- Gibson, B.A.; Kraus, W.L. New Insights into the Molecular and Cellular Functions of Poly(ADP-Ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424.
- Kraus, W.L. PARPs and ADP-Ribosylation: 50 Years … and Counting. Mol. Cell 2015, 58, 902–910.
- Cohen, M.S.; Chang, P. Insights into the Biogenesis, Function, and Regulation of ADP-Ribosylation. Nat. Chem. Biol. 2018, 14, 236–243.
- Barkauskaite, E.; Jankevicius, G.; Ahel, I. Structures and Mechanisms of Enzymes Employed in the Synthesis and Degradation of PARP-Dependent Protein ADP-Ribosylation. Mol. Cell 2015, 58, 935–946.
- Gupte, R.; Liu, Z.; Kraus, W.L. PARPs and ADP-Ribosylation: Recent Advances Linking Molecular Functions to Biological Outcomes. Genes Dev. 2017, 31, 101–126.
- Chaudhuri, A.R.; Nussenzweig, A. The Multifaceted Roles of PARP1 in DNA Repair and Chromatin Remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621.
- Hoch, N.C.; Polo, L.M. ADP-Ribosylation: From Molecular Mechanisms to Human Disease. Genet. Mol. Biol. 2020, 43, e20190075.
- Benjamin, R.C.; Gill, D.M. ADP-Ribosylation in Mammalian Cell Ghosts. Dependence of Poly(ADP-Ribose) Synthesis on Strand Breakage in DNA. J. Biol. Chem. 1980, 255, 10493–10501.
- Durkacz, B.W.; Omidiji, O.; Gray, D.A.; Shall, S. (ADP-Ribose)n Participates in DNA Excision Repair. Nature 1980, 283, 593–596.
- Dockery, L.; Gunderson, C.; Moore, K. Rucaparib: The Past, Present, and Future of a Newly Approved PARP Inhibitor for Ovarian Cancer. OncoTargets Ther. 2017, 10, 3029–3037.
- Fong, P.C.; Boss, D.S.; Yap, T.A.; Tutt, A.; Wu, P.; Mergui-Roelvink, M.; Mortimer, P.; Swaisland, H.; Lau, A.; O’Connor, M.J.; et al. Inhibition of Poly(ADP-Ribose) Polymerase in Tumors from BRCA Mutation Carriers. N. Engl. J. Med. 2009, 361, 123–134.
- Fong, P.C.; Yap, T.A.; Boss, D.S.; Carden, C.P.; Mergui-Roelvink, M.; Gourley, C.; De Greve, J.; Lubinski, J.; Shanley, S.; Messiou, C.; et al. Poly(ADP)-Ribose Polymerase Inhibition: Frequent Durable Responses in BRCA Carrier Ovarian Cancer Correlating With Platinum-Free Interval. J. Clin. Oncol. 2010, 28, 2512–2519.
- Lin, K.K.; Harrell, M.I.; Oza, A.M.; Oaknin, A.; Ray-Coquard, I.; Tinker, A.V.; Helman, E.; Radke, M.R.; Say, C.; Vo, L.-T.; et al. BRCA Reversion Mutations in Circulating Tumor DNA Predict Primary and Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma. Cancer Discov. 2019, 9, 210–219.
- Tuli, R.; Shiao, S.L.; Nissen, N.; Tighiouart, M.; Kim, S.; Osipov, A.; Bryant, M.; Ristow, L.; Placencio-Hickok, V.; Hoffman, D.; et al. A Phase 1 Study of Veliparib, a PARP-1/2 Inhibitor, with Gemcitabine and Radiotherapy in Locally Advanced Pancreatic Cancer. EBioMedicine 2019, 40, 375–381.
- O’Connor, M.J. Targeting the DNA Damage Response in Cancer. Mol. Cell 2015, 60, 547–560.
- Hassa, P.O. The Diverse Biological Roles of Mammalian PARPS, a Small but Powerful Family of Poly-ADP-Ribose Polymerases. Front. Biosci. 2008, 13, 3046.
- Cho, C.C.; Chien, C.Y.; Chiu, Y.C.; Lin, M.H.; Hsu, C.H. Structural and Biochemical Evidence Supporting Poly ADP-Ribosylation in the Bacterium Deinococcus Radiodurans. Nat. Commun. 2019, 10, 14.
- Mennella, M.R.F. The Dichotomy of the Poly(ADP-Ribose) Polymerase-Like Thermozyme from Sulfolobus Solfataricus. Challenges 2018, 9, 5.
- Nishizuka, Y.; Ueda, K.; Nakazawa, K.; Hayaishi, O. Studies on the Polymer of Adenosine Diphosphate Ribose. J. Biol. Chem. 1967, 242, 3164–3171.
- Fujimura, S.; Hasegawa, S.; Shimizu, Y.; Sugimura, T. Polymerization of the Adenosine 5′-Diphosphate-Ribose Moiety of Nicotinamide-Adenine Dinucleotide by Nuclear Enzyme. I. Enzymatic Reactions. Biochim. Biophys Acta 1967, 145, 247–259.
- Ueda, K.; Reeder, R.H.; Honjo, T.; Nishizuka, Y.; Hayaishi, O. Poly Adenosine Diphosphate Ribose Synthesis Associated with Chromatin. Biochem. Biophys. Res. Commun. 1968, 31, 379–385.
- Chambon, P.; Weill, J.D.; Doly, J.; Strosser, M.T.; Mandel, P. On the Formation of a Novel Adenylic Compound by Enzymatic Extracts of Liver Nuclei. Biochem. Biophys. Res. Commun. 1966, 25, 638–643.
- Yamada, M.; Miwa, M.; Sugimura, T. Studies on Poly (Adenosine Diphosphate-Ribose) X. Properties of a Partially Purified Poly (Adenosine Diphosphate-Ribose) Polymerase’. Arch. Biochem. Biophys. 1971, 146, 579–586.
- Juarez-Salinas, H.; Sims, J.L.; Jacobson, M.K. Poly(ADP-Ribose) Levels in Carcinogen-Treated Cells. Nature 1979, 282, 740–741.
- Otake, H.; Miwa, M.; Fujimura, S.; Sugimura, T. Binding of ADP-ribose polymer with histone. J. Biochem. 1969, 65.
- Wang, Z.Q.; Auer, B.; Stingl, L.; Berghammer, H.; Haidacher, D.; Schweiger, M.; Wagner, E.F. Mice Lacking ADPRT and Poly(ADP-Ribosyl)Ation Develop Normally but Are Susceptible to Skin Disease. Genes Dev. 1995, 9, 509–520.
- De Murcia, J.M.; Niedergang, C.; Trucco, C.; Ricoul, M.; Dutrillaux, B.; Mark, M.; Oliver, F.J.; Masson, M.; Dierich, A.; LeMeur, M.; et al. Requirement of Poly(ADP-Ribose) Polymerase in Recovery from DNA Damage in Mice and in Cells. Proc. Natl. Acad. Sci. USA 1997, 94, 7303–7307.
- Masutani, M.; Suzuki, H.; Kamada, N.; Watanabe, M.; Ueda, O.; Nozaki, T.; Jishage, K.-I.; Watanabe, T.; Sugimoto, T.; Nakagama, H.; et al. Poly(ADP-Ribose) Polymerase Gene Disruption Conferred Mice Resistant to Streptozotocin-Induced Diabetes. Proc. Natl. Acad. Sci. USA 1999, 96, 2301–2304.
- D’Amours, D.; Desnoyers, S.; D’Silva, I.; Poirier, G.G. Poly(ADP-Ribosyl)Ation Reactions in the Regulation of Nuclear Functions. Biochem. J. 1999, 342, 249–268.
- Schreiber, V.; Dantzer, F.; Ame, J.-C.; de Murcia, G. Poly(ADP-Ribose): Novel Functions for an Old Molecule. Nat. Rev. Mol. Cell Biol. 2006, 7, 517–528.
- Hakmé, A.; Wong, H.; Dantzer, F.; Schreiber, V. The Expanding Field of Poly(ADP-ribosyl)Ation Reactions. EMBO Rep. 2008, 9, 1094–1100.
- Langelier, M.-F.; Servent, K.M.; Rogers, E.E.; Pascal, J.M. A Third Zinc-Binding Domain of Human Poly(ADP-Ribose) Polymerase-1 Coordinates DNA-Dependent Enzyme Activation. J. Biol. Chem. 2008, 283, 4105–4114.
- Langelier, M.-F.; Ruhl, D.D.; Planck, J.L.; Kraus, W.L.; Pascal, J.M. The Zn3 Domain of Human Poly(ADP-Ribose) Polymerase-1 (PARP-1) Functions in Both DNA-Dependent Poly(ADP-Ribose) Synthesis Activity and Chromatin Compaction*. J. Biol. Chem. 2010, 285, 18877–18887.
- Langelier, M.-F.; Planck, J.L.; Roy, S.; Pascal, J.M. Structural Basis for DNA Damage-Dependent Poly(ADP-Ribosyl)Ation by Human PARP-1. Science 2012, 336, 728–732.
- Amé, J.-C.; Rolli, V.; Schreiber, V.; Niedergang, C.; Apiou, F.; Decker, P.; Muller, S.; Höger, T.; Murcia, J.M.; de Murcia, G. PARP-2, a Novel Mammalian DNA Damage-Dependent Poly(ADP-Ribose) Polymerase. J. Biol. Chem. 1999, 274, 17860–17868.
- Léger, K.; Bär, D.; Savić, N.; Santoro, R.; Hottiger, M.O. ARTD2 Activity Is Stimulated by RNA. Nucleic Acids Res. 2014, 42, 5072–5082.
- Brady, P.N.; Goel, A.; Johnson, M.A. Poly(ADP-Ribose) Polymerases in Host-Pathogen Interactions, Inflammation, and Immunity. Microbiol. Mol. Biol. Rev. 2018, 83.
- Aravind, L.; Koonin, E.V. SAP—A Putative DNA-Binding Motif Involved in Chromosomal Organization. Trends Biochem. Sci. 2000, 25, 112–114.
- Song, J.; Keppler, B.D.; Wise, R.R.; Bent, A.F. PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses. PLoS Genet. 2015, 11, e1005200.
- Rissel, D.; Peiter, E. Poly(ADP-Ribose) Polymerases in Plants and Their Human Counterparts: Parallels and Peculiarities. Int. J. Mol. Sci. 2019, 20, 1638.
- Okubo, S.; Hara, F.; Tsuchida, Y.; Shimotakahara, S.; Suzuki, S.; Hatanaka, H.; Yokoyama, S.; Tanaka, H.; Yasuda, H.; Shindo, H. NMR Structure of the N-Terminal Domain of SUMO Ligase PIAS1 and Its Interaction with Tumor Suppressor P53 and A/T-Rich DNA Oligomers. J. Biol. Chem. 2004, 279, 31455–31461.
- Riccio, A.A.; Cingolani, G.; Pascal, J.M. PARP-2 Domain Requirements for DNA Damage-Dependent Activation and Localization to Sites of DNA Damage. Nucleic Acids Res. 2016, 44, 1691–1702.
- The PyMOL Molecular Graphics System; Schrödinger, LLC: New York, NY, USA, 2021.
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539.
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641.
- Ruf, A.; de Murcia, J.M.; de Murcia, G.; Schulz, G.E. Structure of the Catalytic Fragment of Poly(ADP-Ribose) Polymerase from Chicken. Proc. Natl. Acad. Sci. USA 1996, 93, 7481–7485.
- Ruf, A.; de Murcia, G.; Schulz, G.E. Inhibitor and NAD + Binding to Poly(ADP-Ribose) Polymerase As Derived from Crystal Structures and Homology Modeling †, ‡. Biochemistry 1998, 37, 3893–3900.
- Ruf, A.; Rolli, V.; de Murcia, G.; Schulz, G.E. The Mechanism of the Elongation and Branching Reaction of Poly(ADP-Ribose) Polymerase as Derived from Crystal Structures and Mutagenesis. J. Mol. Biol. 1998, 278, 57–65.
- Oliver, A.W.; Amé, J.-C.; Roe, S.M.; Good, V.; de Murcia, G.; Pearl, L.H. Crystal Structure of the Catalytic Fragment of Murine Poly(ADP-Ribose) Polymerase-2. Nucleic Acids Res. 2004, 32, 456–464.
- Kinoshita, T.; Nakanishi, I.; Warizaya, M.; Iwashita, A.; Kido, Y.; Hattori, K.; Fujii, T. Inhibitor-Induced Structural Change of the Active Site of Human Poly(ADP-Ribose) Polymerase. FEBS Lett. 2004, 556, 43–46.
- Karlberg, T.; Hammarström, M.; Schütz, P.; Svensson, L.; Schüler, H. Crystal Structure of the Catalytic Domain of Human PARP2 in Complex with PARP Inhibitor ABT-888. Biochemistry 2010, 49, 1056–1058.
- Lehtiö, L.; Jemth, A.-S.; Collins, R.; Loseva, O.; Johansson, A.; Markova, N.; Hammarström, M.; Flores, A.; Holmberg-Schiavone, L.; Weigelt, J.; et al. Structural Basis for Inhibitor Specificity in Human Poly(ADP-Ribose) Polymerase-3 †. J. Med. Chem. 2009, 52, 3108–3111.
- Bilokapic, S.; Suskiewicz, M.J.; Ahel, I.; Halic, M. Bridging of DNA Breaks Activates PARP2–HPF1 to Modify Chromatin. Nature 2020, 585, 609–613.
- Caldecott, K.W. Single-Strand Break Repair and Genetic Disease. Nat. Rev. Genet. 2008, 9, 619–631.
- White, R.R.; Vijg, J. Do DNA Double-Strand Breaks Drive Aging? Mol. Cell 2016, 63, 729–738.
- Silva, E.; Ideker, T. Transcriptional Responses to DNA Damage. DNA Repair 2019, 79, 40–49.
- Langelier, M.-F.; Riccio, A.A.; Pascal, J.M. PARP-2 and PARP-3 Are Selectively Activated by 5′ Phosphorylated DNA Breaks Through an Allosteric Regulatory Mechanism Shared with PARP-1. Nucleic Acids Res. 2014, 42, 7762–7775.
- Hiom, K. Coping with DNA Double Strand Breaks. DNA Repair 2010, 9, 1256–1263.
- Caldecott, K.W. XRCC1 Protein; Form and Function. DNA Repair 2019, 81, 102664.
- Das, B.B.; Huang, S.N.; Murai, J.; Rehman, I.; Amé, J.-C.; Sengupta, S.; Das, S.K.; Majumdar, P.; Zhang, H.; Biard, D.; et al. PARP1–TDP1 Coupling for the Repair of Topoisomerase I–Induced DNA Damage. Nucleic Acids Res. 2014, 42, 4435–4449.
- Spagnolo, L.; Barbeau, J.; Curtin, N.J.; Morris, E.P.; Pearl, L.H. Visualization of a DNA-PK/PARP1 Complex. Nucleic Acids Res. 2012, 40, 4168–4177.
- Roth, D.B. V(D)J Recombination: Mechanism, Errors, and Fidelity. In Mobile DNA III; John Wiley & Sons: Hoboken, NJ, USA, 2015.
- Haince, J.-F.; Kozlov, S.; Dawson, V.L.; Dawson, T.M.; Hendzel, M.J.; Lavin, M.F.; Poirier, G.G. Ataxia Telangiectasia Mutated (ATM) Signaling Network Is Modulated by a Novel Poly(ADP-Ribose)-Dependent Pathway in the Early Response to DNA-Damaging Agents. J. Biol. Chem. 2007, 282, 16441–16453.
- Aguilar-Quesada, R.; Muñoz-Gámez, J.; Martín-Oliva, D.; Peralta, A.; Valenzuela, M.T.; Matínez-Romero, R.; Quiles-Pérez, R.; Murcia, J.; de Murcia, G.; de Almodóvar, M.; et al. Interaction between ATM and PARP-1 in Response to DNA Damage and Sensitization of ATM Deficient Cells Through PARP Inhibition. BMC Mol. Biol. 2007, 8, 29.
- Haince, J.-F.; McDonald, D.; Rodrigue, A.; Déry, U.; Masson, J.-Y.; Hendzel, M.J.; Poirier, G.G. PARP1-Dependent Kinetics of Recruitment of MRE11 and NBS1 Proteins to Multiple DNA Damage Sites. J. Biol. Chem. 2008, 283, 1197–1208.
- Grabarz, A.; Barascu, A.; Guirouilh-Barbat, J.; Lopez, B.S. Initiation of DNA Double Strand Break Repair: Signaling and Single-Stranded Resection Dictate the Choice Between Homologous Recombination, Non-Homologous End-Joining and Alternative End-Joining. Am. J. Cancer Res. 2012, 2, 249–268.
- Langelier, M.-F.; Planck, J.L.; Roy, S.; Pascal, J.M. Crystal Structures of Poly(ADP-Ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: Structural and Functional Insights into DNA-Dependent PARP-1 Activity. J. Biol. Chem. 2011, 286, 10690–10701.
- Ali, A.A.E.; Timinszky, G.; Arribas-Bosacoma, R.; Kozlowski, M.; Hassa, P.O.; Hassler, M.; Ladurner, A.G.; Pearl, L.H.; Oliver, A.W. The Zinc-Finger Domains of PARP1 Cooperate to Recognize DNA Strand Breaks. Nat. Struct. Mol. Biol. 2012, 19, 685–692.
- Eustermann, S.; Wu, W.-F.; Langelier, M.-F.; Yang, J.-C.; Easton, L.E.; Riccio, A.A.; Pascal, J.M.; Neuhaus, D. Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1. Mol. Cell 2015, 60, 742–754.
- Obaji, E.; Haikarainen, T.; Lehtiö, L. Structural Basis for DNA Break Recognition by ARTD2/PARP2. Nucleic Acids Res. 2018, 46, 12154–12165.
- Eustermann, S.; Videler, H.; Yang, J.-C.; Cole, P.T.; Gruszka, D.; Veprintsev, D.; Neuhaus, D. The DNA-Binding Domain of Human PARP-1 Interacts with DNA Single-Strand Breaks as a Monomer through Its Second Zinc Finger. J. Mol. Biol. 2011, 407, 149–170.
- Trucco, C.; Flatter, E.; Fribourg, S.; de Murcia, G.; Ménissier-de Murcia, J. Mutations in the Amino-Terminal Domain of the Human Poly(ADP-Ribose) Polymerase That Affect Its Catalytic Activity but Not Its DNA Binding Capacity. FEBS Lett. 1996, 399, 313–316.
- Rulten, S.L.; Fisher, A.E.O.; Robert, I.; Zuma, M.C.; Rouleau, M.; Ju, L.; Poirier, G.; Reina-San-Martin, B.; Caldecott, K.W. PARP-3 and APLF Function Together to Accelerate Nonhomologous End-Joining. Mol. Cell 2011, 41, 33–45.
- Zarkovic, G.; Belousova, E.A.; Talhaoui, I.; Saint-Pierre, C.; Kutuzov, M.M.; Matkarimov, B.T.; Biard, D.; Gasparutto, D.; Lavrik, O.I.; Ishchenko, A.A. Characterization of DNA ADP-Ribosyltransferase Activities of PARP2 and PARP3: New Insights into DNA ADP-Ribosylation. Nucleic Acids Res. 2018, 46, 2417–2431.
- Liu, L.; Kong, M.; Gassman, N.R.; Freudenthal, B.D.; Prasad, R.; Zhen, S.; Watkins, S.C.; Wilson, S.H.; Van Houten, B. PARP1 Changes from Three-Dimensional DNA Damage Searching to One-Dimensional Diffusion after Auto-PARylation or in the Presence of APE1. Nucleic Acids Res. 2017, 45, 12834–12847.
- Milo, R.; Jorgensen, P.; Moran, U.; Weber, G.; Springer, M. BioNumbers—the Database of Key Numbers in Molecular and Cell Biology. Nucleic Acids Res. 2010, 38, D750–D753.
- Huber, M.D.; Gerace, L. The Size-Wise Nucleus: Nuclear Volume Control in Eukaryotes. J. Cell. Biol. 2007, 179, 583–584.
- Sukhanova, M.V.; Abrakhi, S.; Joshi, V.; Pastre, D.; Kutuzov, M.M.; Anarbaev, R.O.; Curmi, P.A.; Hamon, L.; Lavrik, O.I. Single Molecule Detection of PARP1 and PARP2 Interaction with DNA Strand Breaks and Their Poly(ADP-Ribosyl)Ation Using High-Resolution AFM Imaging. Nucleic Acids Res. 2016, 44, e60.
- Sukhanova, M.V.; Hamon, L.; Kutuzov, M.M.; Joshi, V.; Abrakhi, S.; Dobra, I.; Curmi, P.A.; Pastre, D.; Lavrik, O.I. A Single-Molecule Atomic Force Microscopy Study of PARP1 and PARP2 Recognition of Base Excision Repair DNA Intermediates. J. Mol. Biol. 2019, 431, 2655–2673.
- Rudolph, J.; Mahadevan, J.; Dyer, P.; Luger, K. Poly(ADP-Ribose) Polymerase 1 Searches DNA via a ‘Monkey Bar’ Mechanism. eLife 2018, 7, e37818.
- Rudolph, J.; Mahadevan, J.; Luger, K. Probing the Conformational Changes Associated with DNA Binding to PARP1. Biochemistry 2020, 59, 2003–2011.
