Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Others
Laxaphycins were first isolated by Moore’s Hawaiian group in 1992 [19,20]. They were extracted from the cyanobacterium Anabaena laxa, collected in mud on the campus of the University of Hawaii.
- laxaphycin
- cyanobacteria
- synthesis
- biosynthesis
- marine natural product
- peptide
1. Cyclic Laxaphycins
Laxaphycins were first isolated by Moore’s Hawaiian group in 1992 [19,20]. They were extracted from the cyanobacterium Anabaena laxa, collected in mud on the campus of the University of Hawaii. After successive fractionation steps, four major laxaphycins A, B, D, and E were isolated from about thirteen other similar peptides. The raw cyclic structures of the four laxaphycins were determined. These four peptides could already be separated into two subfamilies, laxaphycins with 11 residues (lax A, E) and laxaphycins containing 12 residues (lax B, D), each of them containing in their sequences at least 50% of non-ribosomal amino acids. Laxaphycin A is composed of a 3-aminooctanoic acid residue (Aoc), two homoserine (Hse), a E-dehydrobutyrine residue (Dhb), a 4-hydroxyproline (Hyp), and six more standard amino acids with the following sequence (Aoc-Hse-E-Dhb-Hyp-Hse-Phe-Leu-Ile-Ile-Leu-Gly). The two isoleucines are diastereoisomers. Laxaphycin E showed a unique difference with laxaphycin A, the substitution of the 3-aminooctanoic acid (Aoc) by a 3-aminodecanoic acid (Ade). Laxaphycin B contains the non-proteinogenic amino acids 3-aminodecanoic acid (Ade), 3-hydroxyleucine (Hle), 3-hydroxyasparagine (Hasn), and N-methylisoleucine ((N)-MeIle) as well as six natural amino acids, with the sequence (Ade-Val-Hle-Gln-(N)-MeIle-Hasn-Thr-Pro-Leu-Thr). Laxaphycin D differentiated from laxaphycin B by a 3-aminooctanoic acid that replaces the 3-aminodecanoic acid.
It is only in 1997 that Bonnard et al. identified the complete structure and assigned the absolute stereochemistry of the amino acids constituting the laxaphycins A and B sequence [21]. The two peptides were extracted from an assemblage of the marine cyanobacteria Lyngbya majuscula (L. majuscula) and Anabaena torulosa (A. torulosa), collected in Moorea (French Polynesia), but repeated harvesting at Moorea island highlighted A. torulosa as the only producer of laxaphycins. Different experiments including mass spectrometry and NMR confirmed the established gross structure, whereas combining the previous results with Marfey’s analysis gave access to the stereochemistry of all amino acids. Thus, for laxaphycin A, 3-aminooctanoic acid, phenylalanine, and leucine in positions 1, 6, and 7, respectively were shown to be of R-configuration. Isoleucine in position 8 is of (2S,3S)-configuration while isoleucine in position 9 has a (2R,3S) configuration, the other amino acids of the sequence having a classical S-configuration. It is worth noting that for laxaphycin A, hydrophobic residues are located on the same side of the peptide. In laxaphycin B the stereochemistry of 3-hydroxyleucine in position 3 was (2S,3S) while 3-hydroxyleucine in position 5 was (2R,3S). The 3-hydroxyasparagine was stereochemically (2R,3R). An alternation of L and D stereochemistry can be seen along the peptide sequence, with (3R)-Ade1, (2R,3S)-3-OH-Leu5, (2R,3R)-3-OH-Asn8 and d-Leu11, and the hydrophobic/hydrophilic character of amino acids is also alternated within the sequence. A further study of the biological activities of laxaphycins produced by cyanobacteria was carried out by the same group in 2007, giving rise at the same time to two new laxaphycins named laxaphycins B2 and B3 (Figure 1) [22]. Laxaphycin B2 differs from laxaphycin B only by the replacement of (2R,3S)-3-hydroxyleucine to d-leucine in position 5. Laxaphycin B3 contains a (2S,4R)-4-hydroxyproline in position 10 instead of the proline found in laxaphycin B. In these two new laxaphycins, the 3-hydroxyleucine in position 3 is of the (2R,3S) configuration whereas the one described in laxaphycin B is of the (2S,3S) configuration.
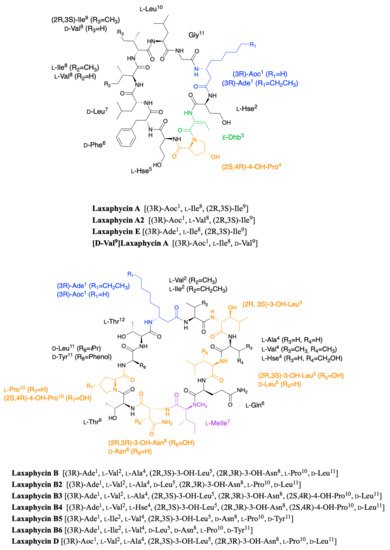
Figure 1. Structure of cyclic laxaphycins A and B.
In order to further study the ecological role of laxaphycin B and its potential in the discovery of new anti-cancer agents, a total synthesis was achieved in 2013 highlighting a structural misassignment for the absolute configuration of one amino acid [23,24]. Indeed, the comparison of synthetic and natural laxaphycin B showed that laxaphycin B had a (2R,3S)-3-hydroxyleucine in position 3 whereas it had been described with a (2S,3S) configuration (Figure 1).
About a decade later, new analogues of the laxaphycin family were reported, having little difference with the already known laxaphycins [25,26]. Laxaphycins A2 and B4 were discovered in an extract of the cyanobacterium Hormothamnion enteromorphoides collected in the Gulf of Mexico. Laxaphycin A2 contains an l-valine in position 8 while laxaphycin A contains an l-isoleucine (Figure 1) [25]. Laxaphycin B4 is analogous to laxaphycin B3, with a homoserine in position 4 replacing the alanine (Figure 1) [25]. Two new peptides laxaphycins B5 and B6 isolated from a freshwater cyanobacterium, Phormidium sp., UIC 10484 completed this series [26]. Laxaphycin B5 is derived from laxaphycin B and contains isoleucine instead of valine in position 2, valine in position 4 instead of alanine, asparagine instead of 3-hydroxyasparagine in position 8, and tyrosine in position 11 instead of leucine. Similarly, laxaphycin B6 combines the same four previous modifications observed in laxaphycin B5 and a d-leucine in position 5 that made it one of the more distant compound from laxaphycin B. It is worth to note that all the observed modifications are made with maintenance of stereochemistry and that the amino acids are often isosteric. The tyrosine and leucine in position 11 are both d-configuration, as are asparagine and 3-hydroxyasparagine in position 8.
One year later, an extract of A. torulosa collected in Moorea (French Polynesia) revealed two cyclic laxaphycins-A type peptides, one of which was the previously described laxaphycin A2 [27]. The other, named [d-Val9]laxaphycin A, showed a very similar structure, but NMR and MSMS analyses suggested the presence of d-valine in position 9 instead of (2R,3S)-isoleucine, while keeping (2S,3S)-isoleucine in position 8 (Figure 1). These two new cyclic laxaphycin A were not found in other samples of Anabaena sp. collected at different locations in the lagoon of Moorea [28]. The production of these peptides cannot yet be explained. Peptides with close structures have been found in other cyanobacteria such as Hormothamnion enteromorphoides [29] or Lyngbya confervoides [30]. A horizontal gene transfer between cyanobacteria could be an explanation for the presence of these peptides within different cyanobacteria strains [31].
2. Acyclic Laxaphycins
To complete this set of compounds, three acyclic laxaphycin A type peptides were discovered [27]. Acyclolaxaphycin A is equivalent to laxaphycin A opened between (3R)-Aoc in position 1 and glycine in position 11. NMR and MS spectra of [des-(Gly11)]acyclolaxaphycin A show a loss of glycine 11. The [des-(Leu10-Gly11)]acyclolaxaphycin A corresponds to acyclolaxaphycin A having lost glycine in position 11 and leucine in position 10 (Figure 2). Like their cyclic structural analogues, laxaphycin A2 and [d-Val9]laxaphycin A, these new acyclolaxaphycins A were not found in samples of Anabaena sp. collected by Bonnard et al. for their study on the toxicity of cyanobacterial blooms, thus highlighting that laxaphycin production might be seasonally affected or dependent on the environmental conditions [28].
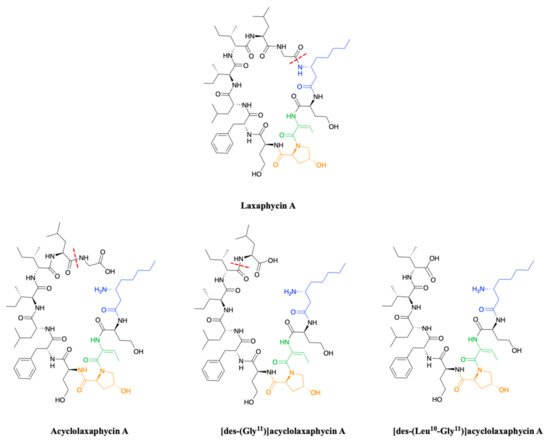
Figure 2. Structure of acyclic laxaphycins A compared to laxaphycin A. Amino acid deletion leading to the next acyclic laxaphycin A are indicated by red marks.
In 2015, upon examination of an extract of A. torulosa, Bornancin et al. also found two acyclic laxaphycins, B and B3, that differ from their cyclic homologs by an m/z value 18 amu higher [32]. Furthermore, both peptides responded positively to the ninhydrin test, confirming an uncapped N-terminus. After fragmentation by MS-MS, the peptide is proposed to be open between 3-hydroxyleucine in position 3 and alanine in position 4. Stereochemistry is preserved for all amino acids between the cyclic and acyclic versions (Figure 3). These two new acyclolaxaphycins B were not found in other samples of Anabaena sp. collected at different locations in the lagoon of Moorea, as in the case of the new laxaphycins A mentioned above [28]. Recently, two acyclolaxaphycins were isolated from the sea hare Stylocheilus striatus that feeds on the cyanobacterium A. torulosa, producing laxaphycins [33]. High resolution mass spectrometry (HRMS) analysis allowed to propose [des-(Ala4-Hle5)]acyclolaxaphycins B and B3 that result from the loss of the two successive alanine and 3-hydroxyleucine in positions 4 and 5 (Figure 3). NMR analyses sustained this proposal due to the absence of correlations between alanine and a 3-hydroxyleucine. These two compounds undergo a spontaneous cyclization of the N-terminal glutamine to form a pyroglutamate, resulting in [des-(Ala4-Hle5)]acyclolaxaphycin B3 and B1211, respectively (Figure 3) [34].
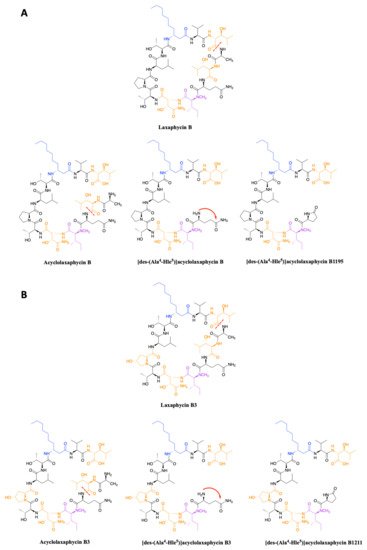
Figure 3. (A) Structure of acyclic laxaphycins B compared to cyclic laxaphycin B; (B) Structure of acyclic laxaphycins B3 compared to cyclic laxaphycin B3. Positions of ring opening or structural modifications leading to the next acyclic laxaphycins are designated by red marks.
This entry is adapted from the peer-reviewed paper 10.3390/md19090473
This entry is offline, you can click here to edit this entry!
